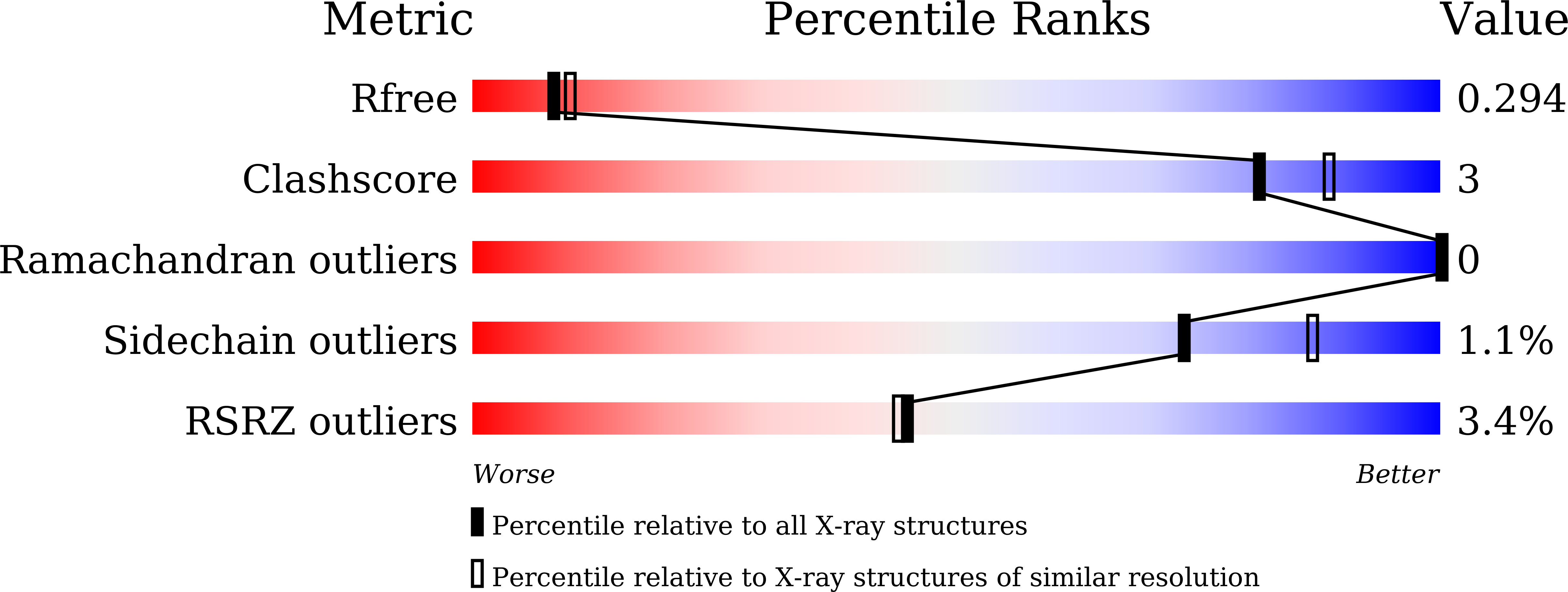
Structures of the NDP-pyranose mutase belonging to glycosyltransferase family 75 reveal residues important for Mn 2+ coordination and substrate binding.
Du, X., Chu, X., Liu, N., Jia, X., Peng, H., Xiao, Y., Liu, L., Yu, H., Li, F., He, C.(2023) J Biol Chem 299: 102903-102903
- PubMed: 36642179
- DOI: https://doi.org/10.1016/j.jbc.2023.102903
- Primary Citation of Related Structures:
7XPR, 7XPS, 7XPT, 7XPU, 7XPV, 8HL8 - PubMed Abstract:
Members of glycosyltransferase family 75 (GT75) not only reversibly catalyze the autoglycosylation of a conserved arginine residue with specific NDP-sugars but also exhibit NDP-pyranose mutase activity that reversibly converts specific NDP-pyranose to NDP-furanose. The latter activity provides valuable NDP-furanosyl donors for glycosyltransferases and requires a divalent cation as a cofactor instead of FAD used by UDP-D-galactopyranose mutase. However, details of the mechanism for NDP-pyranose mutase activity are not clear. Here we report the first crystal structures of GT75 family NDP-pyranose mutases. The novel structures of GT75 member MtdL in complex with Mn 2+ and GDP, GDP-D-glucopyranose, GDP-L-fucopyranose, GDP-L-fucofuranose, respectively, combined with site-directed mutagenesis studies, reveal key residues involved in Mn 2+ coordination, substrate binding, and catalytic reactions. We also provide a possible catalytic mechanism for this unique type of NDP-pyranose mutase. Taken together, our results highlight key elements of an enzyme family important for furanose biosynthesis.
Organizational Affiliation:
Anhui Key Laboratory of Modern Biomanufacturing and School of Life Sciences, Anhui University, Hefei, Anhui, China.