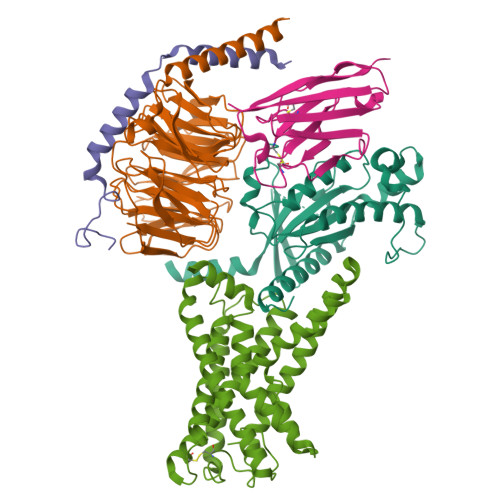
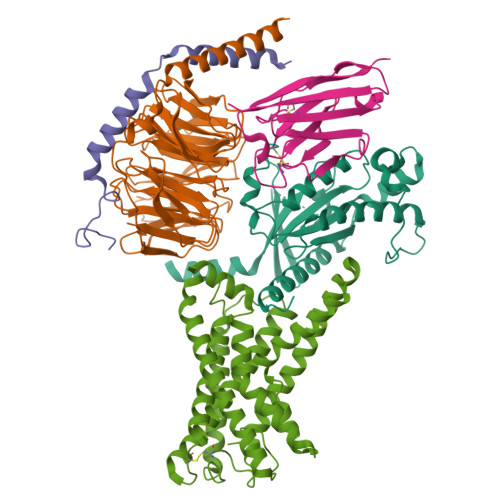
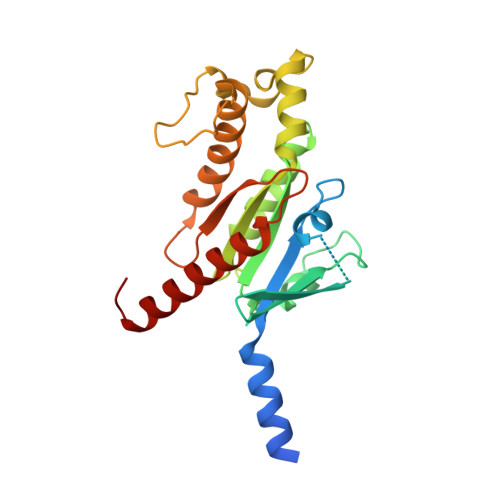
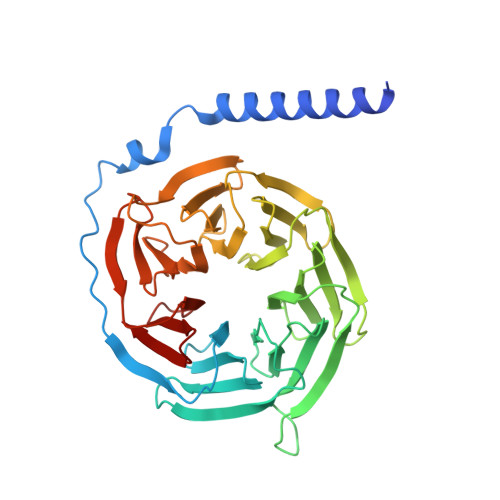
Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Qu, X., Qiu, N., Wang, M., Zhang, B., Du, J., Zhong, Z., Xu, W., Chu, X., Ma, L., Yi, C., Han, S., Shui, W., Zhao, Q., Wu, B.(2022) Nature 604: 779-785
- PubMed: 35418679
- DOI: https://doi.org/10.1038/s41586-022-04580-w
- Primary Citation of Related Structures:
7WU2, 7WU3, 7WU4, 7WU5 - PubMed Abstract:
Adhesion G protein-coupled receptors (aGPCRs) are essential for a variety of physiological processes such as immune responses, organ development, cellular communication, proliferation and homeostasis 1-7 . An intrinsic manner of activation that involves a tethered agonist in the N-terminal region of the receptor has been proposed for the aGPCRs 8,9 , but its molecular mechanism remains elusive. Here we report the G protein-bound structures of ADGRD1 and ADGRF1, which exhibit many unique features with regard to the tethered agonism. The stalk region that proceeds the first transmembrane helix acts as the tethered agonist by forming extensive interactions with the transmembrane domain; these interactions are mostly conserved in ADGRD1 and ADGRF1, suggesting that a common stalk-transmembrane domain interaction pattern is shared by members of the aGPCR family. A similar stalk binding mode is observed in the structure of autoproteolysis-deficient ADGRF1, supporting a cleavage-independent manner of receptor activation. The stalk-induced activation is facilitated by a cascade of inter-helix interaction cores that are conserved in positions but show sequence variability in these two aGPCRs. Furthermore, the intracellular region of ADGRF1 contains a specific lipid-binding site, which proves to be functionally important and may serve as the recognition site for the previously discovered endogenous ADGRF1 ligand synaptamide. These findings highlight the diversity and complexity of the signal transduction mechanisms of the aGPCRs.
Organizational Affiliation:
CAS Key Laboratory of Receptor Research, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China.