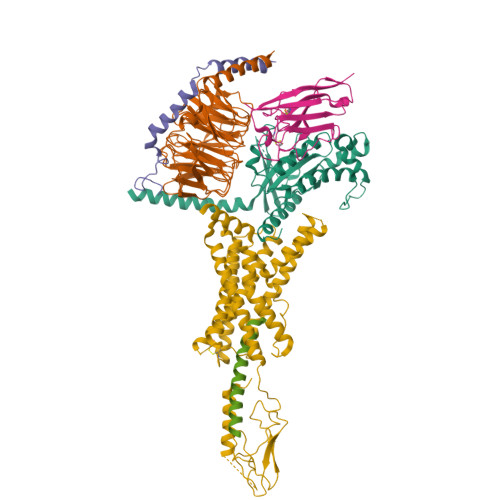
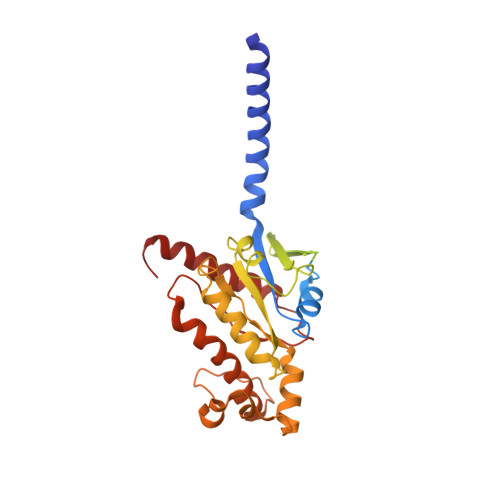
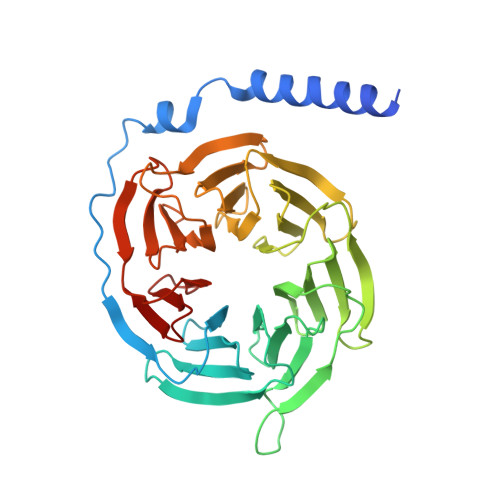
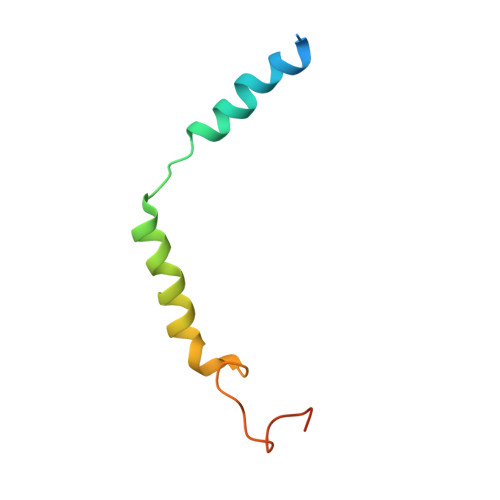
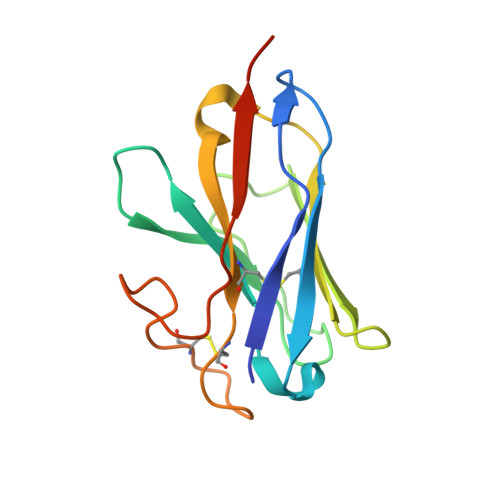
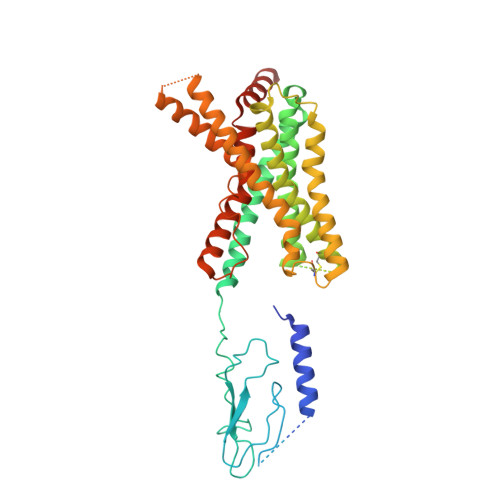
Endogenous ligand recognition and structural transition of a human PTH receptor.
Kobayashi, K., Kawakami, K., Kusakizako, T., Miyauchi, H., Tomita, A., Kobayashi, K., Shihoya, W., Yamashita, K., Nishizawa, T., Kato, H.E., Inoue, A., Nureki, O.(2022) Mol Cell 82: 3468-3483.e5
- PubMed: 35932760
- DOI: https://doi.org/10.1016/j.molcel.2022.07.003
- Primary Citation of Related Structures:
7VVJ, 7VVK, 7VVL, 7VVM, 7VVN, 7VVO - PubMed Abstract:
Endogenous parathyroid hormone (PTH) and PTH-related peptide (PTHrP) bind to the parathyroid hormone receptor 1 (PTH1R) and activate the stimulatory G-protein (Gs) signaling pathway. Intriguingly, the two ligands have distinct signaling and physiological properties: PTH evokes prolonged Gs activation, whereas PTHrP evokes transient Gs activation with reduced bone-resorption effects. The distinct molecular actions are ascribed to the differences in ligand recognition and dissociation kinetics. Here, we report cryoelectron microscopic structures of six forms of the human PTH1R-Gs complex in the presence of PTH or PTHrP at resolutions of 2.8 -4.1 Å. A comparison of the PTH-bound and PTHrP-bound structures reveals distinct ligand-receptor interactions underlying the ligand affinity and selectivity. Furthermore, five distinct PTH-bound structures, combined with computational analyses, provide insights into the unique and complex process of ligand dissociation from the receptor and shed light on the distinct durations of signaling induced by PTH and PTHrP.
Organizational Affiliation:
Department of Biological Sciences, Graduate School of Science, the University of Tokyo, Bunkyo, Tokyo 113-0033, Japan.