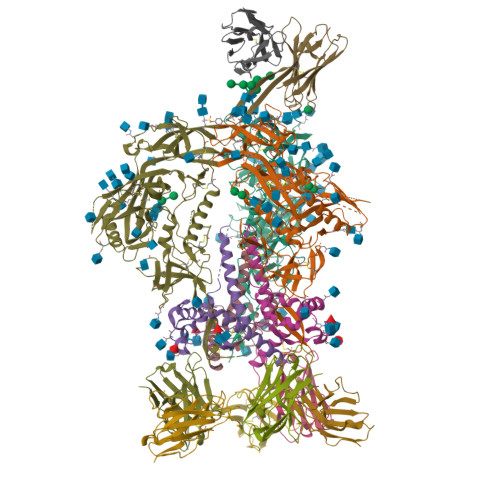
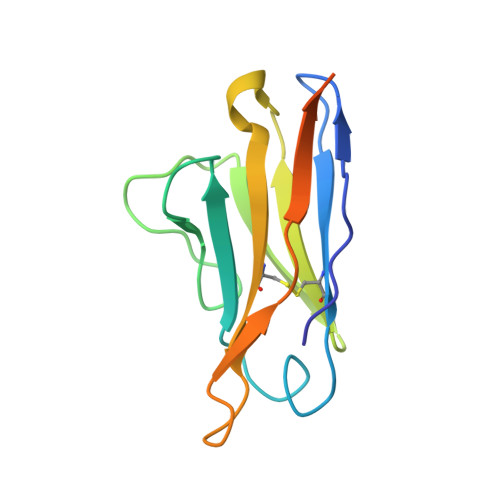
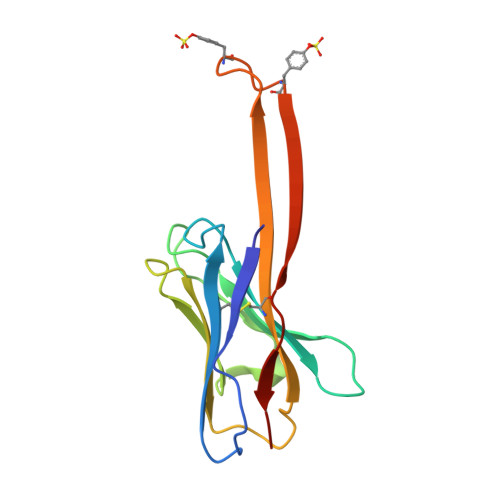
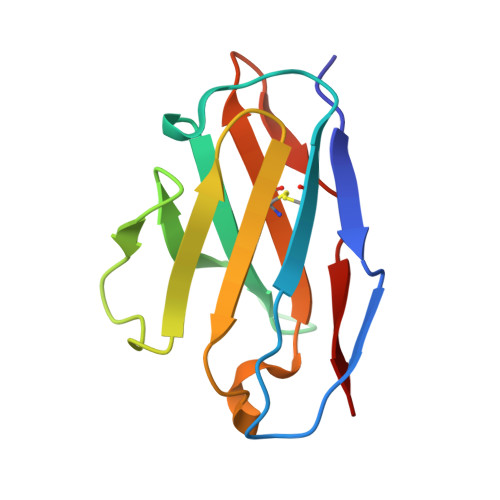
NAG Query on NAG Download Ideal Coordinates CCD File AB [auth C]
AB [auth C],
2-acetamido-2-deoxy-beta-D-glucopyranose 8 H15 N O6