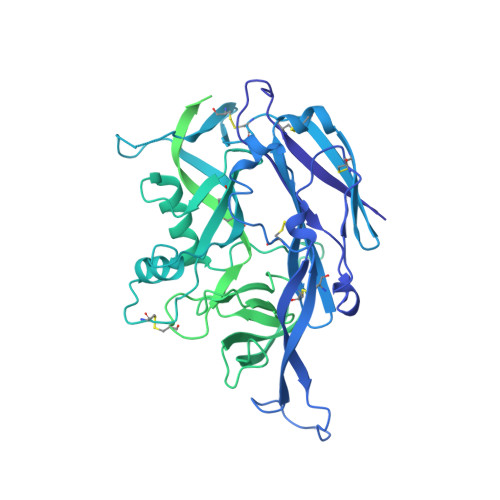
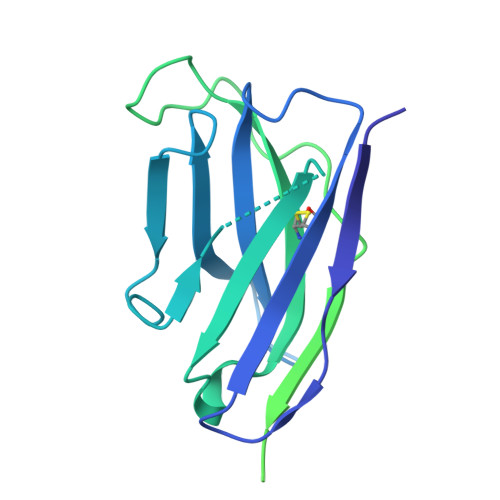
Structural and mechanistic basis of neutralization by a pan-hantavirus protective antibody.
Mittler, E., Serris, A., Esterman, E.S., Florez, C., Polanco, L.C., O'Brien, C.M., Slough, M.M., Tynell, J., Groning, R., Sun, Y., Abelson, D.M., Wec, A.Z., Haslwanter, D., Keller, M., Ye, C., Bakken, R.R., Jangra, R.K., Dye, J.M., Ahlm, C., Rappazzo, C.G., Ulrich, R.G., Zeitlin, L., Geoghegan, J.C., Bradfute, S.B., Sidoli, S., Forsell, M.N.E., Strandin, T., Rey, F.A., Herbert, A.S., Walker, L.M., Chandran, K., Guardado-Calvo, P.(2023) Sci Transl Med 15: eadg1855-eadg1855
- PubMed: 37315110
- DOI: https://doi.org/10.1126/scitranslmed.adg1855
- Primary Citation of Related Structures:
7QQB - PubMed Abstract:
Emerging rodent-borne hantaviruses cause severe diseases in humans with no approved vaccines or therapeutics. We recently isolated a monoclonal broadly neutralizing antibody (nAb) from a Puumala virus-experienced human donor. Here, we report its structure bound to its target, the Gn/Gc glycoprotein heterodimer comprising the viral fusion complex. The structure explains the broad activity of the nAb: It recognizes conserved Gc fusion loop sequences and the main chain of variable Gn sequences, thereby straddling the Gn/Gc heterodimer and locking it in its prefusion conformation. We show that the nAb's accelerated dissociation from the divergent Andes virus Gn/Gc at endosomal acidic pH limits its potency against this highly lethal virus and correct this liability by engineering an optimized variant that sets a benchmark as a candidate pan-hantavirus therapeutic.
Organizational Affiliation:
Department of Microbiology and Immunology, Albert Einstein College of Medicine, Bronx, NY 10461, USA.