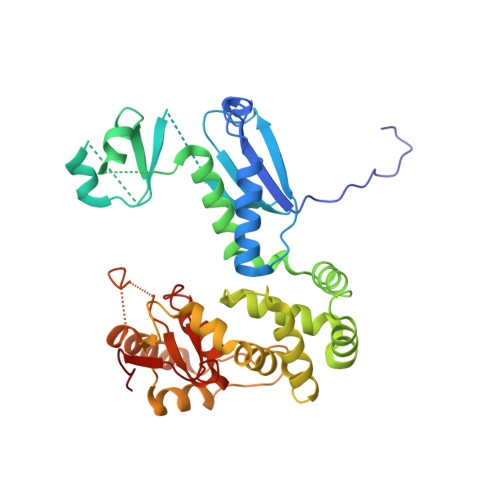
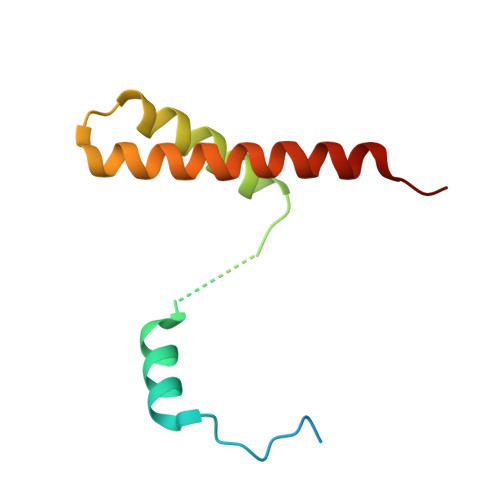
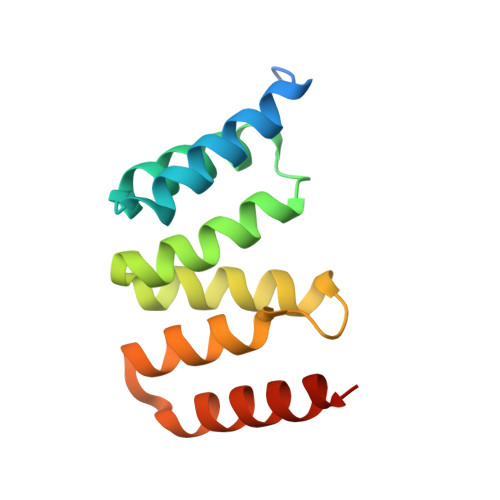Direct interaction of a chaperone-bound type III secretion substrate with the export gate.
Gilzer, D., Schreiner, M., Niemann, H.H.(2022) Nat Commun 13: 2858-2858
- PubMed: 35654781
- DOI: https://doi.org/10.1038/s41467-022-30487-1
- Primary Citation of Related Structures:
7QIH, 7QII, 7QIJ - PubMed Abstract:
Several gram-negative bacteria employ type III secretion systems (T3SS) to inject effector proteins into eukaryotic host cells directly from the bacterial cytoplasm. The export gate SctV (YscV in Yersinia) binds substrate:chaperone complexes such as YscX:YscY, which are essential for formation of a functional T3SS. Here, we present structures of the YscX:YscY complex alone and bound to nonameric YscV. YscX binds its chaperone YscY at two distinct sites, resembling the heterotrimeric complex of the T3SS needle subunit with its chaperone and co-chaperone. In the ternary complex the YscX N-terminus, which mediates YscX secretion, occupies a binding site within one YscV that is also used by flagellar chaperones, suggesting the interaction's importance for substrate recognition. The YscX C-terminus inserts between protomers of the YscV ring where the stalk protein binds to couple YscV to the T3SS ATPase. This primary YscV-YscX interaction is essential for the formation of a secretion-competent T3SS.
Organizational Affiliation:
Department of Chemistry, Bielefeld University, Universitaetstrasse 25, 33615, Bielefeld, Germany.