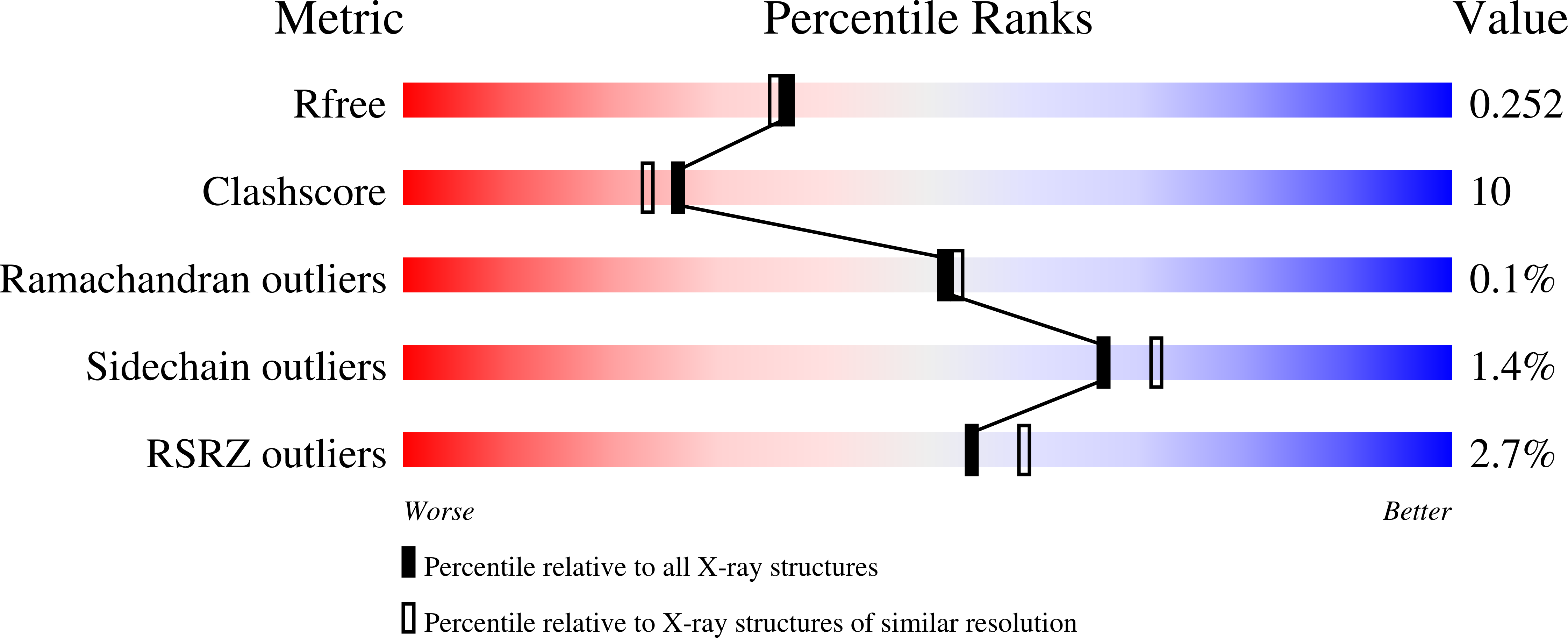
Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Neun, S., Brear, P., Campbell, E., Tryfona, T., El Omari, K., Wagner, A., Dupree, P., Hyvonen, M., Hollfelder, F.(2022) Nat Chem Biol 18: 1096-1103
- PubMed: 35799064
- DOI: https://doi.org/10.1038/s41589-022-01071-x
- Primary Citation of Related Structures:
7QE1, 7QE2, 7QEA, 7QEE, 7QEF, 7QG4 - PubMed Abstract:
The abundance of recorded protein sequence data stands in contrast to the small number of experimentally verified functional annotation. Here we screened a million-membered metagenomic library at ultrahigh throughput in microfluidic droplets for β-glucuronidase activity. We identified SN243, a genuine β-glucuronidase with little homology to previously studied enzymes of this type, as a glycoside hydrolase 3 family member. This glycoside hydrolase family contains only one recently added β-glucuronidase, showing that a functional metagenomic approach can shed light on assignments that are currently 'unpredictable' by bioinformatics. Kinetic analyses of SN243 characterized it as a promiscuous catalyst and structural analysis suggests regions of divergence from homologous glycoside hydrolase 3 members creating a wide-open active site. With a screening throughput of >10 7 library members per day, picolitre-volume microfluidic droplets enable functional assignments that complement current enzyme database dictionaries and provide bridgeheads for the annotation of unexplored sequence space.
Organizational Affiliation:
Department of Biochemistry, University of Cambridge, Cambridge, UK.