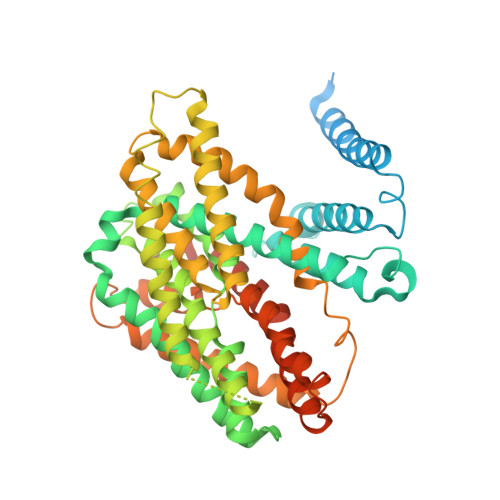
Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Matsuoka, R., Fudim, R., Jung, S., Zhang, C., Bazzone, A., Chatzikyriakidou, Y., Robinson, C.V., Nomura, N., Iwata, S., Landreh, M., Orellana, L., Beckstein, O., Drew, D.(2022) Nat Struct Mol Biol 29: 108-120
- PubMed: 35173351
- DOI: https://doi.org/10.1038/s41594-022-00738-2
- Primary Citation of Related Structures:
7P1I, 7P1J, 7P1K - PubMed Abstract:
The Na + /H + exchanger SLC9B2, also known as NHA2, correlates with the long-sought-after Na + /Li + exchanger linked to the pathogenesis of diabetes mellitus and essential hypertension in humans. Despite the functional importance of NHA2, structural information and the molecular basis for its ion-exchange mechanism have been lacking. Here we report the cryo-EM structures of bison NHA2 in detergent and in nanodiscs, at 3.0 and 3.5 Å resolution, respectively. The bison NHA2 structure, together with solid-state membrane-based electrophysiology, establishes the molecular basis for electroneutral ion exchange. NHA2 consists of 14 transmembrane (TM) segments, rather than the 13 TMs previously observed in mammalian Na + /H + exchangers (NHEs) and related bacterial antiporters. The additional N-terminal helix in NHA2 forms a unique homodimer interface with a large intracellular gap between the protomers, which closes in the presence of phosphoinositol lipids. We propose that the additional N-terminal helix has evolved as a lipid-mediated remodeling switch for the regulation of NHA2 activity.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Stockholm University, Stockholm, Sweden.