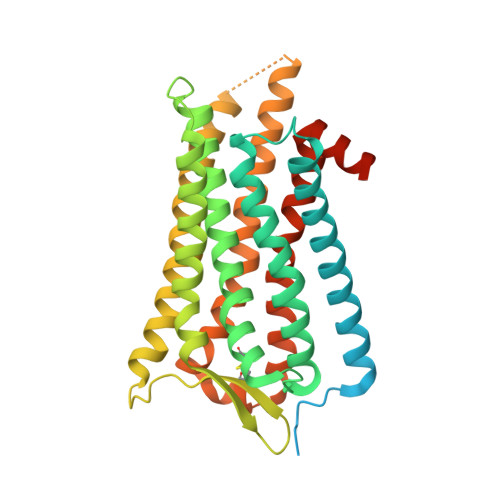
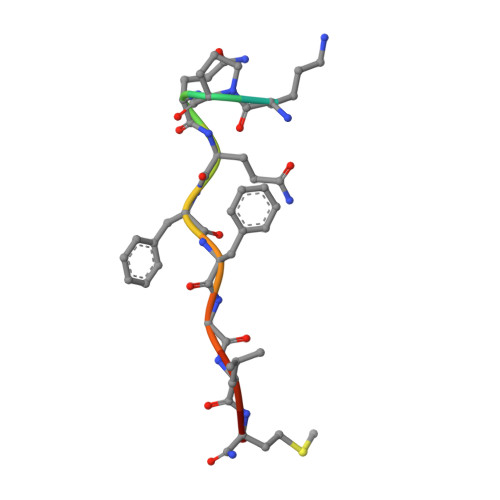
Structures of neurokinin 1 receptor in complex with G q and G s proteins reveal substance P binding mode and unique activation features.
Thom, C., Ehrenmann, J., Vacca, S., Waltenspuhl, Y., Schoppe, J., Medalia, O., Pluckthun, A.(2021) Sci Adv 7: eabk2872-eabk2872
- PubMed: 34878828
- DOI: https://doi.org/10.1126/sciadv.abk2872
- Primary Citation of Related Structures:
7P00, 7P02 - PubMed Abstract:
The neurokinin 1 receptor (NK 1 R) is involved in inflammation and pain transmission. This pathophysiologically important G protein–coupled receptor is predominantly activated by its cognate agonist substance P (SP) but also by the closely related neurokinins A and B. Here, we report cryo–electron microscopy structures of SP-bound NK 1 R in complex with its primary downstream signal mediators, G q and G s . Our structures reveal how a polar network at the extracellular, solvent-exposed receptor surface shapes the orthosteric pocket and that NK 1 R adopts a noncanonical active-state conformation with an interface for G protein binding, which is distinct from previously reported structures. Detailed comparisons with antagonist-bound NK 1 R crystal structures reveal that insurmountable antagonists induce a distinct and long-lasting receptor conformation that sterically blocks SP binding. Together, our structures provide important structural insights into ligand and G protein promiscuity, the lack of basal signaling, and agonist- and antagonist-induced conformations in the neurokinin receptor family.
- Department of Biochemistry, University of Zürich, Winterthurerstrasse 190, CH-8057 Zürich, Switzerland.
Organizational Affiliation: