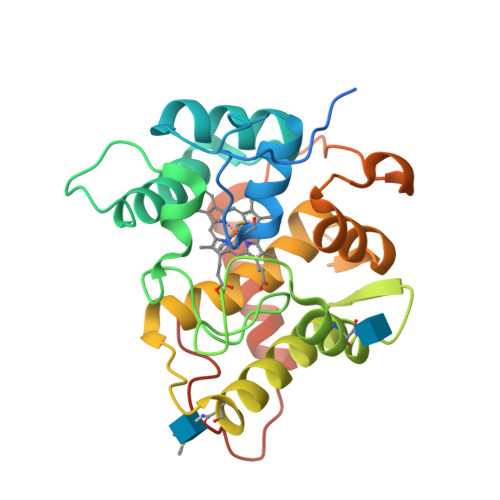
Structural and biochemical studies enlighten the unspecific peroxygenase from Hypoxylon sp. EC38 as an efficient oxidative biocatalyst.
Rotilio, L., Swoboda, A., Ebner, K., Rinnofner, C., Glieder, A., Kroutil, W., Mattevi, A.(2021) ACS Catal 11: 11511-11525
- PubMed: 34540338
- DOI: https://doi.org/10.1021/acscatal.1c03065
- Primary Citation of Related Structures:
7O1R, 7O1X, 7O1Z, 7O2D, 7O2G - PubMed Abstract:
Unspecific peroxygenases (UPO) are glycosylated fungal enzymes that can selectively oxidize C-H bonds. UPOs employ hydrogen peroxide as oxygen donor and reductant. With such an easy-to-handle co-substrate and without the need of a reducing agent, UPOs are emerging as convenient oxidative biocatalysts. Here, an unspecific peroxygenase from Hypoxylon sp. EC38 ( Hsp UPO) was identified in an activity-based screen of six putative peroxygenase enzymes that were heterologously expressed in Pichia pastoris . The enzyme was found to tolerate selected organic solvents such as acetonitrile and acetone. Hsp UPO is a versatile catalyst performing various reactions, such as the oxidation of prim - and sec -alcohols, epoxidations and hydroxylations. Semi-preparative biotransformations were demonstrated for the non-enantioselective oxidation of racemic 1-phenylethanol rac -1b (TON = 13000), giving the product with 88% isolated yield, and the oxidation of indole 6a to give indigo 6b (TON = 2800) with 98% isolated yield. Hsp UPO features a compact and rigid three-dimensional conformation that wraps around the heme and defines a funnel-shaped tunnel that leads to the heme iron from the protein surface. The tunnel extends along a distance of about 12 Å with a fairly constant diameter in its innermost segment. Its surface comprises both hydrophobic and hydrophilic groups for dealing with small-to-medium size substrates of variable polarities. The structural investigation of several protein-ligand complexes revealed that the active site of Hsp UPO is accessible to molecules of varying bulkiness and polarity with minimal or no conformational changes, explaining the relatively broad substrate scope of the enzyme. With its convenient expression system, robust operational properties, relatively small size, well-defined structural features, and diverse reaction scope, Hsp UPO is an exploitable candidate for peroxygenase-based biocatalysis.
Organizational Affiliation:
Department of Biology and Biotechnology, University of Pavia, via Ferrata 9, 27100 Pavia, Italy.