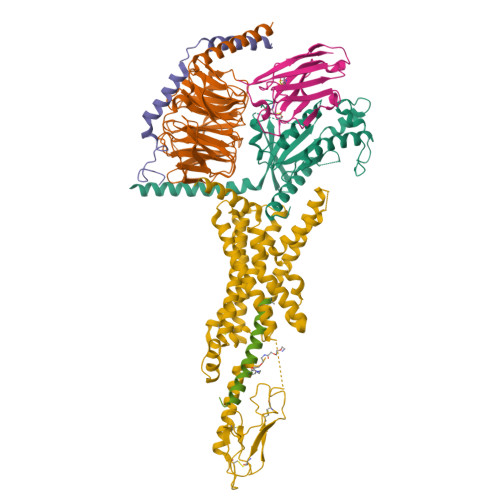
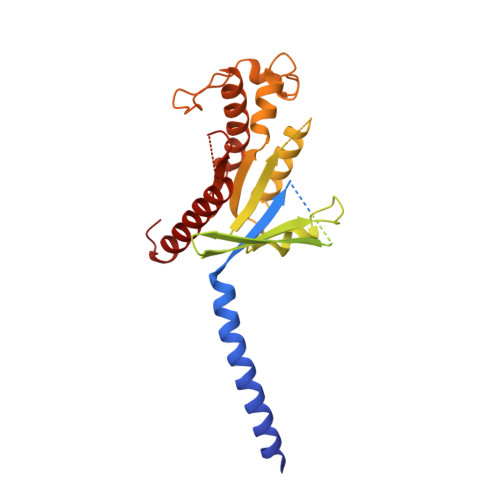
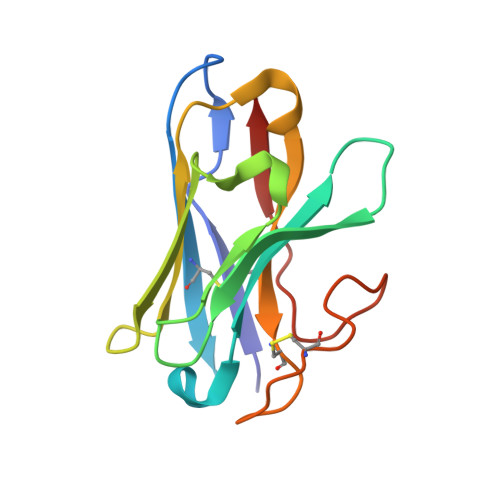
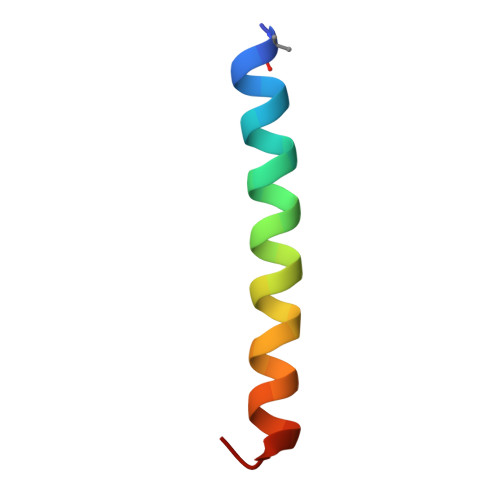
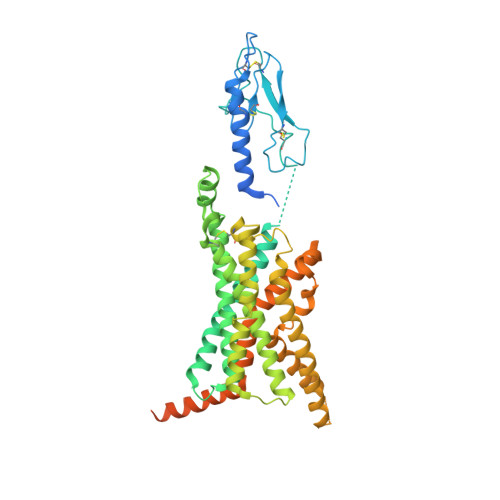
Structure and dynamics of semaglutide- and taspoglutide-bound GLP-1R-Gs complexes.
Zhang, X., Belousoff, M.J., Liang, Y.L., Danev, R., Sexton, P.M., Wootten, D.(2021) Cell Rep 36: 109374-109374
- PubMed: 34260945
- DOI: https://doi.org/10.1016/j.celrep.2021.109374
- Primary Citation of Related Structures:
7KI0, 7KI1 - PubMed Abstract:
The glucagon-like peptide-1 receptor (GLP-1R) regulates insulin secretion, carbohydrate metabolism, and appetite and is an important target for treatment of type 2 diabetes and obesity. Multiple GLP-1R agonists have entered into clinical trials, with some, such as semaglutide, progressing to approval. Others, including taspoglutide, failed due to the high incidence of side effects or insufficient efficacy. GLP-1R agonists have a broad spectrum of signaling profiles, but molecular understanding is limited by a lack of structural information on how different agonists engage with the GLP-1R. Here, we report cryoelectron microscopy (cryo-EM) structures and cryo-EM 3D variability analysis of semaglutide- and taspoglutide-bound GLP-1R-Gs protein complexes. These reveal similar peptide interactions to GLP-1 but different motions within the receptor and bound peptides, providing insights into the molecular determinants of GLP-1R peptide engagement.
Organizational Affiliation:
Drug Discovery Biology, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville 3052, VIC, Australia; ARC Centre for Cryo-electron Microscopy of Membrane Proteins, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville 3052, VIC, Australia.