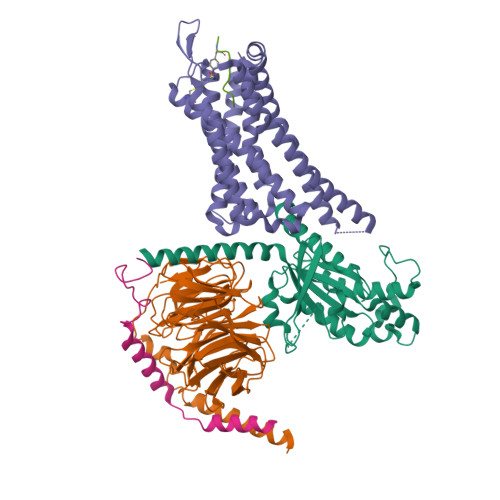
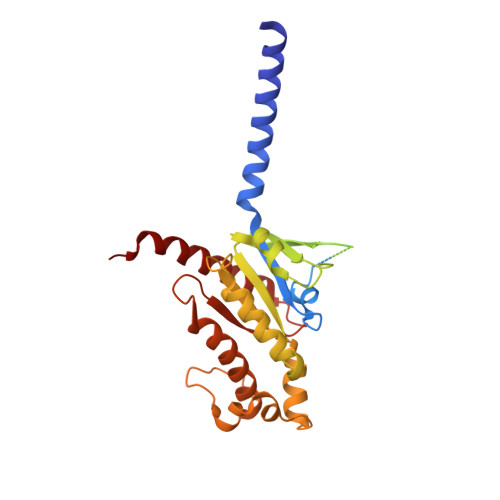
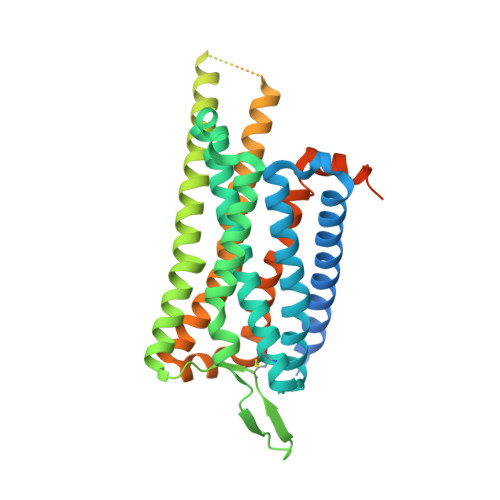
Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor
Liu, Q., Yang, D., Zhuang, Y., Croll, T.I., Cai, X., Dai, A., He, X., Duan, J., Yin, W., Ye, C., Zhou, F., Wu, B., Zhao, Q., Xu, H.E., Wang, M.W., Jiang, Y.(2021) Nat Chem Biol 17: 1238-1244