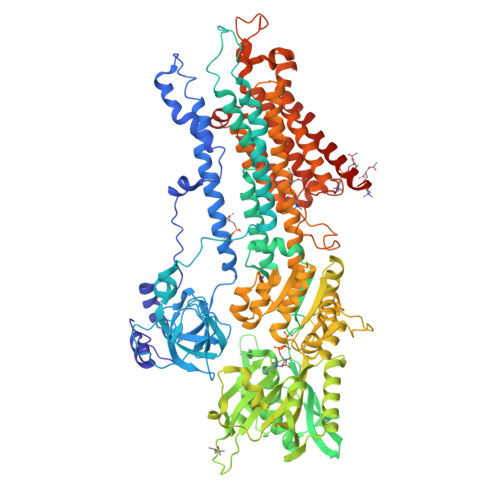
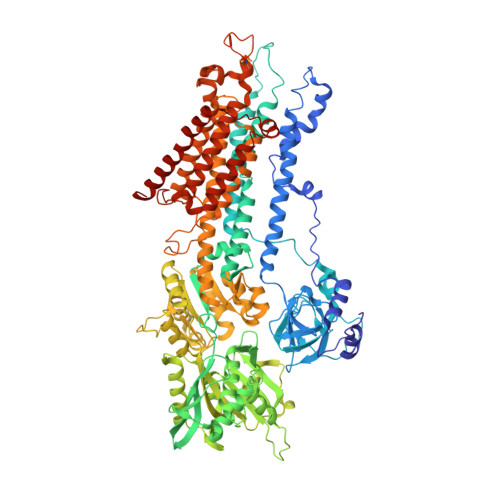
What ATP binding does to the Ca2+pump and how nonproductive phosphoryl transfer is prevented in the absence of Ca2.
Kabashima, Y., Ogawa, H., Nakajima, R., Toyoshima, C.(2020) Proc Natl Acad Sci U S A 117: 18448-18458
- PubMed: 32675243
- DOI: https://doi.org/10.1073/pnas.2006027117
- Primary Citation of Related Structures:
7BT2 - PubMed Abstract:
Under physiological conditions, most Ca 2+ -ATPase (SERCA) molecules bind ATP before binding the Ca 2+ transported. SERCA has a high affinity for ATP even in the absence of Ca 2+ , and ATP accelerates Ca 2+ binding at pH values lower than 7, where SERCA is in the E2 state with low-affinity Ca 2+ -binding sites. Here we describe the crystal structure of SERCA2a, the isoform predominant in cardiac muscle, in the E2·ATP state at 3.0-Å resolution. In the crystal structure, the arrangement of the cytoplasmic domains is distinctly different from that in canonical E2. The A-domain now takes an E1 position, and the N-domain occupies exactly the same position as that in the E1·ATP·2Ca 2+ state relative to the P-domain. As a result, ATP is properly delivered to the phosphorylation site. Yet phosphoryl transfer never takes place without the filling of the two transmembrane Ca 2+ -binding sites. The present crystal structure explains what ATP binding itself does to SERCA and how nonproductive phosphorylation is prevented in E2.
Organizational Affiliation:
Institute for Quantitative Biosciences, The University of Tokyo, 113-0032 Tokyo, Japan.