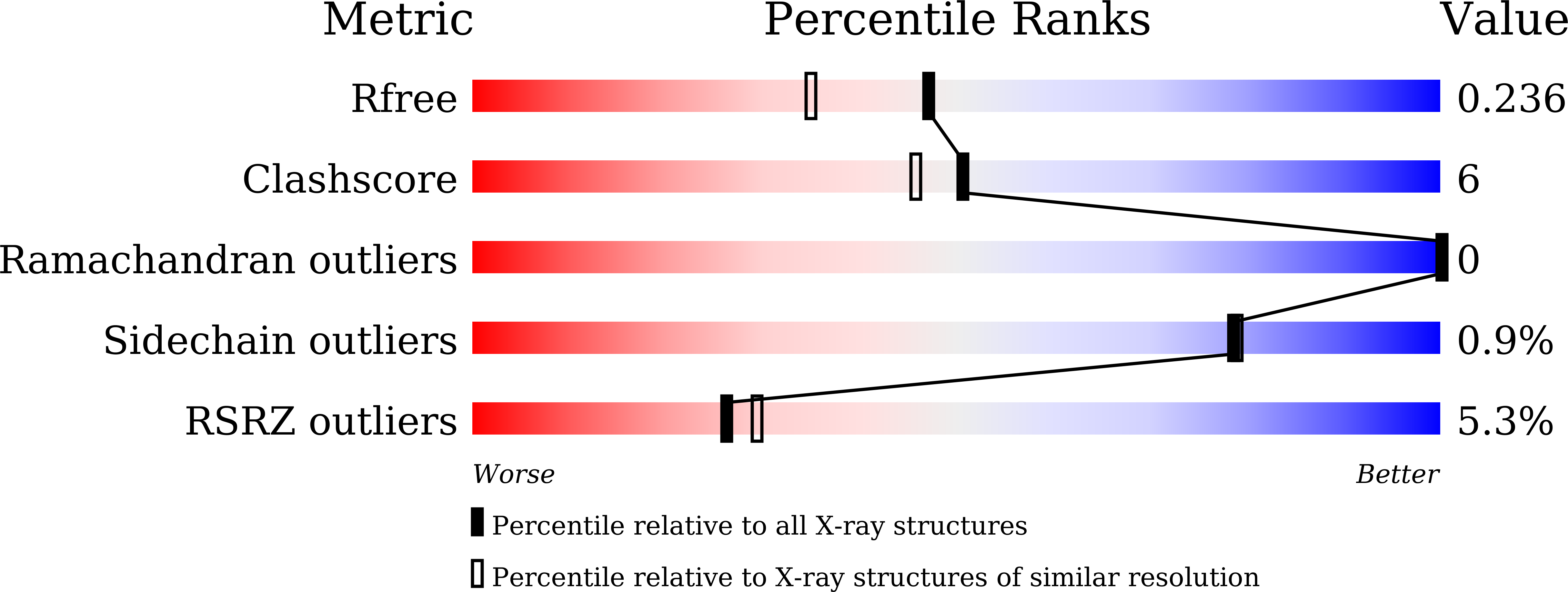
Structural bases of IMiD selectivity that emerges by 5-hydroxythalidomide.
Furihata, H., Yamanaka, S., Honda, T., Miyauchi, Y., Asano, A., Shibata, N., Tanokura, M., Sawasaki, T., Miyakawa, T.(2020) Nat Commun 11: 4578-4578
- PubMed: 32929090
- DOI: https://doi.org/10.1038/s41467-020-18488-4
- Primary Citation of Related Structures:
7BQU, 7BQV - PubMed Abstract:
Thalidomide and its derivatives exert not only therapeutic effects as immunomodulatory drugs (IMiDs) but also adverse effects such as teratogenicity, which are due in part to different C2H2 zinc-finger (ZF) transcription factors, IKZF1 (or IKZF3) and SALL4, respectively. Here, we report the structural bases for the SALL4-specific proteasomal degradation induced by 5-hydroxythalidomide, a primary thalidomide metabolite generated by the enzymatic activity of cytochrome P450 isozymes, through the interaction with cereblon (CRBN). The crystal structure of the metabolite-mediated human SALL4-CRBN complex and mutagenesis studies elucidate the complex formation enhanced by the interaction between CRBN and an additional hydroxy group of (S)-5-hydroxythalidomide and the variation in the second residue of β-hairpin structure that underlies the C2H2 ZF-type neo-morphic substrate (neosubstrate) selectivity of 5-hydroxythalidomide. These findings deepen our understanding of the pharmaceutical action of IMiDs and provide structural evidence that the glue-type E3 ligase modulators cause altered neosubstrate specificities through their metabolism.
Organizational Affiliation:
Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo, 113-8657, Japan.