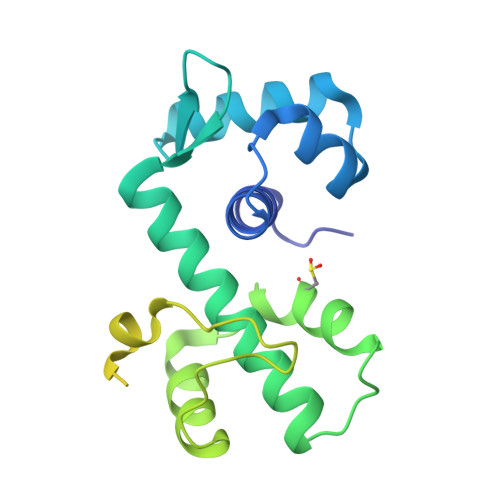
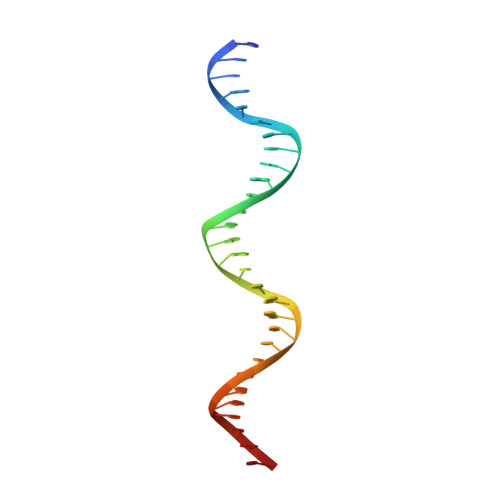
The bacterial iron sensor IdeR recognizes its DNA targets by indirect readout.
Marcos-Torres, F.J., Maurer, D., Juniar, L., Griese, J.J.(2021) Nucleic Acids Res 49: 10120-10135
- PubMed: 34417623
- DOI: https://doi.org/10.1093/nar/gkab711
- Primary Citation of Related Structures:
7B1V, 7B1Y, 7B20, 7B23, 7B24, 7B25 - PubMed Abstract:
The iron-dependent regulator IdeR is the main transcriptional regulator controlling iron homeostasis genes in Actinobacteria, including species from the Corynebacterium, Mycobacterium and Streptomyces genera, as well as the erythromycin-producing bacterium Saccharopolyspora erythraea. Despite being a well-studied transcription factor since the identification of the Diphtheria toxin repressor DtxR three decades ago, the details of how IdeR proteins recognize their highly conserved 19-bp DNA target remain to be elucidated. IdeR makes few direct contacts with DNA bases in its target sequence, and we show here that these contacts are not required for target recognition. The results of our structural and mutational studies support a model wherein IdeR mainly uses an indirect readout mechanism, identifying its targets via the sequence-dependent DNA backbone structure rather than through specific contacts with the DNA bases. Furthermore, we show that IdeR efficiently recognizes a shorter palindromic sequence corresponding to a half binding site as compared to the full 19-bp target previously reported, expanding the number of potential target genes controlled by IdeR proteins.
Organizational Affiliation:
Department of Cell and Molecular Biology, Uppsala University, SE-751 24 Uppsala, Sweden.