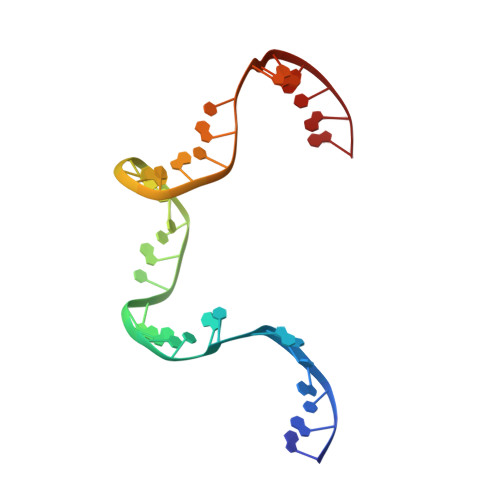
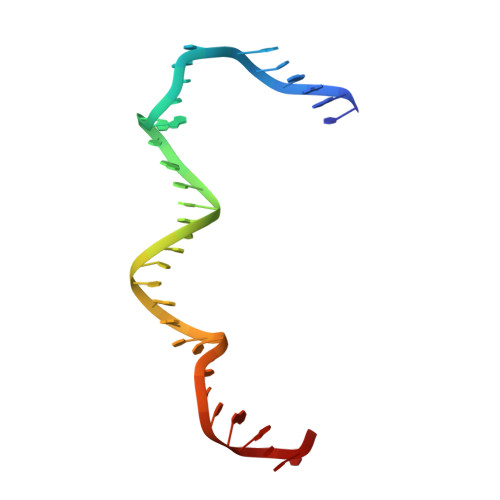
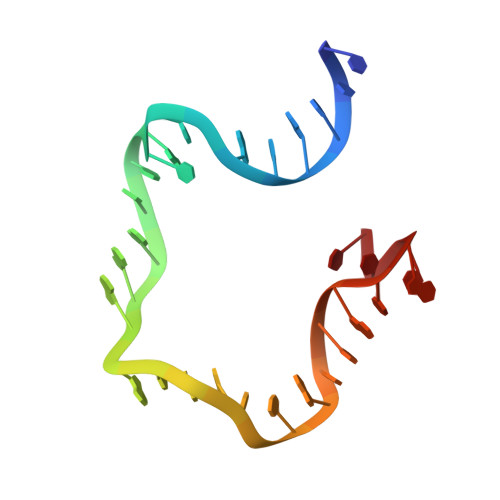
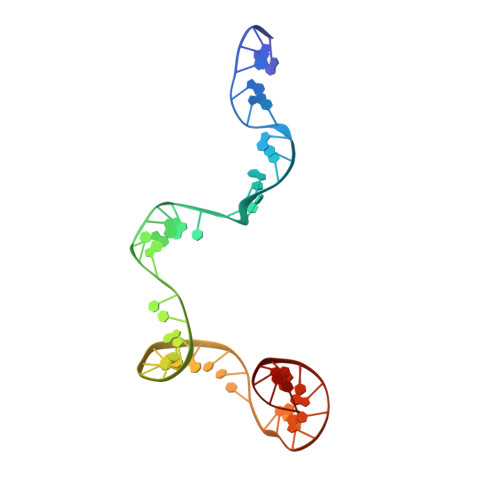
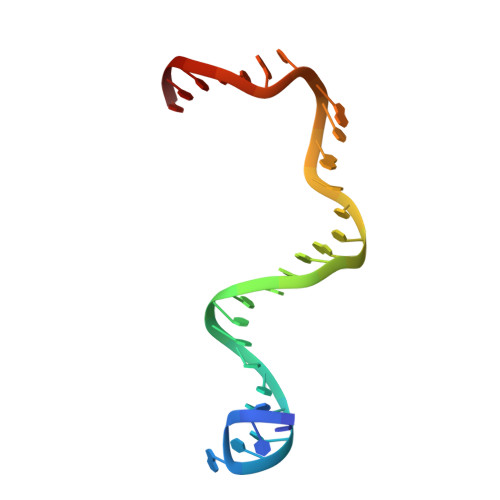
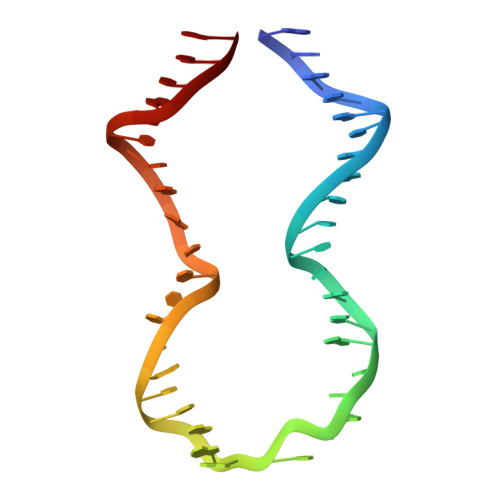
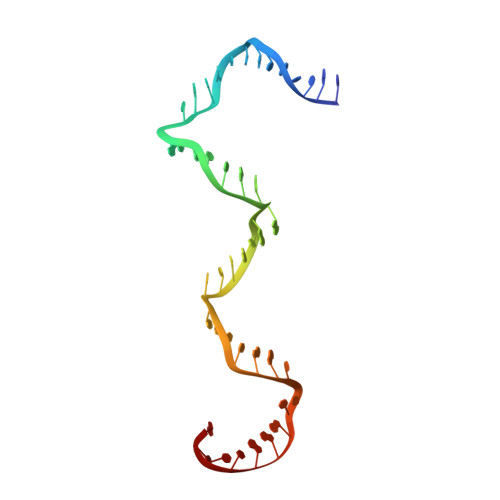
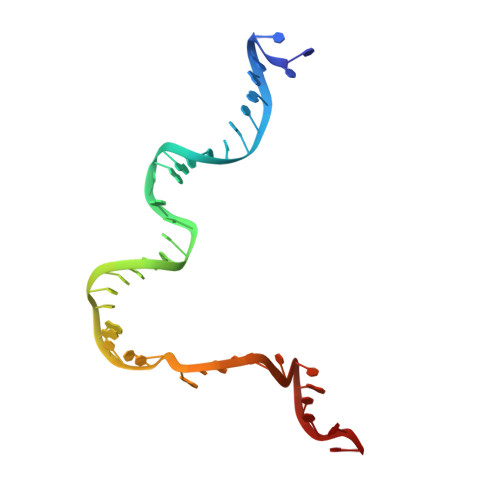
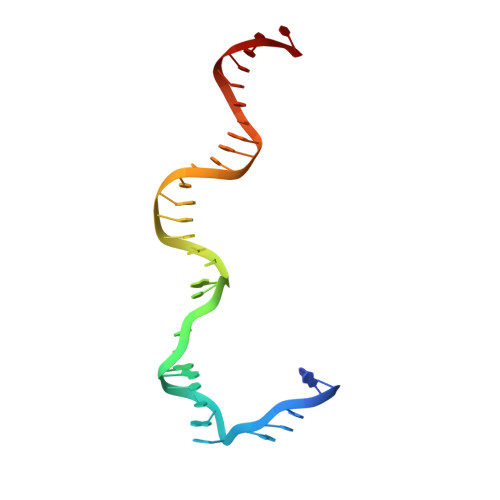
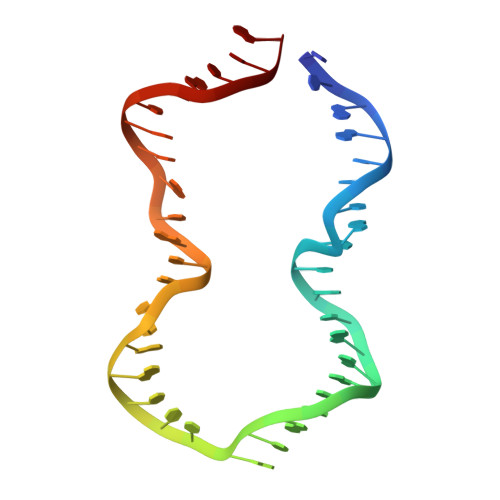
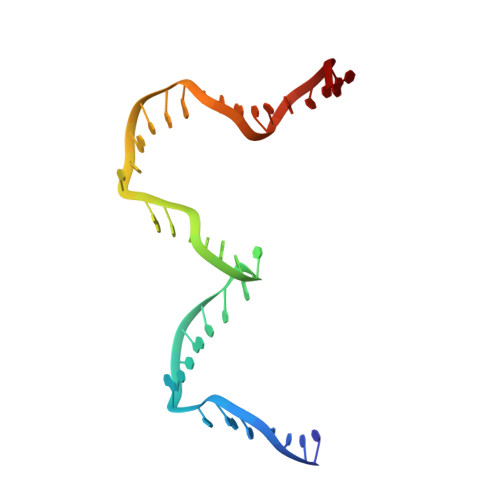
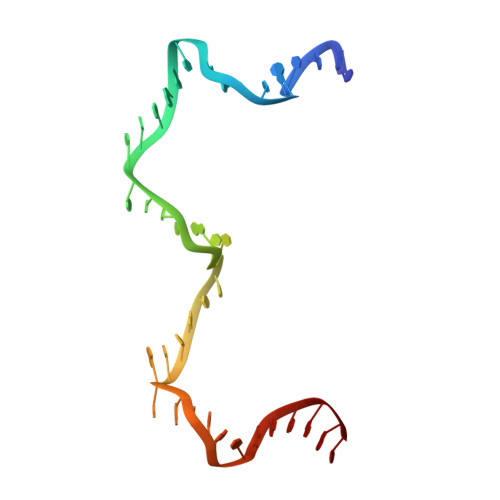
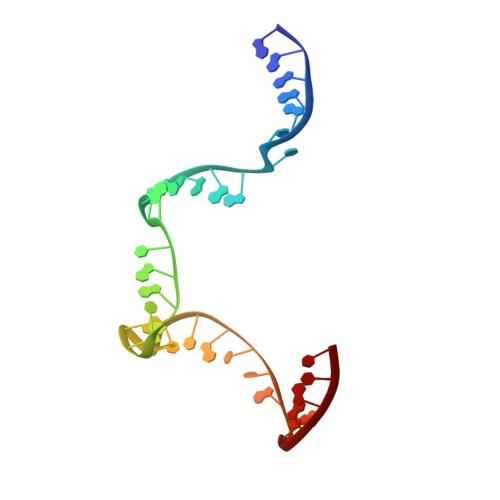
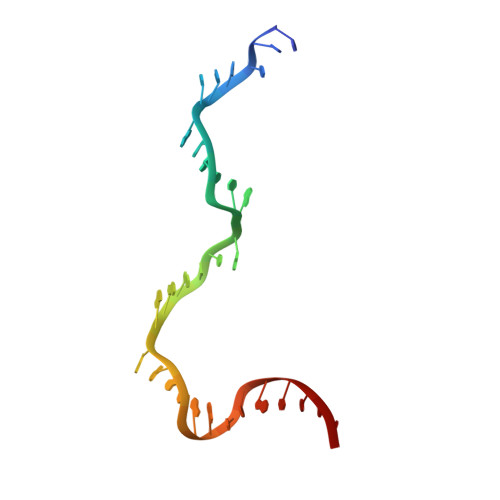
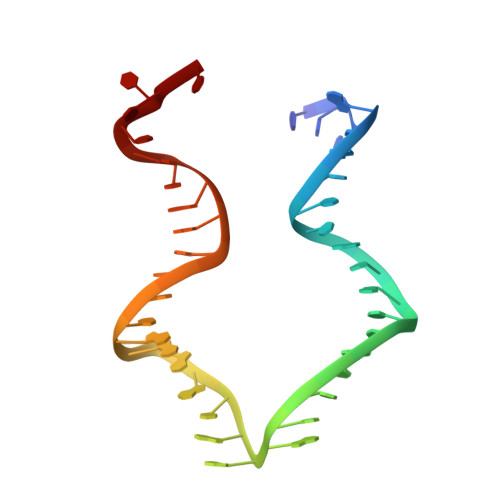
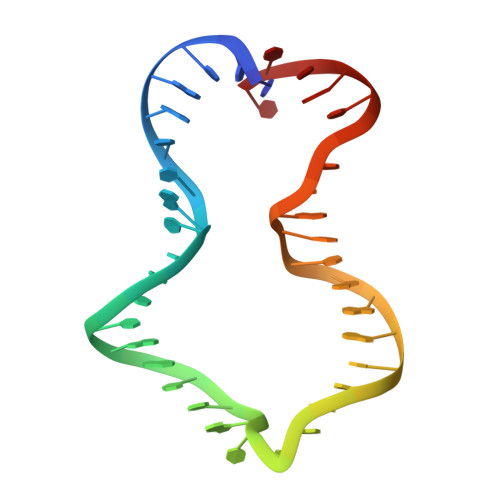
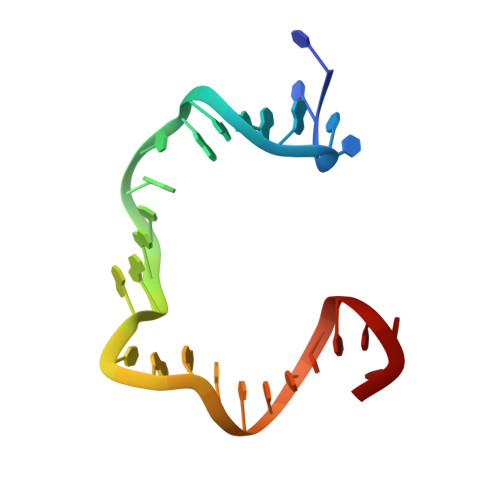
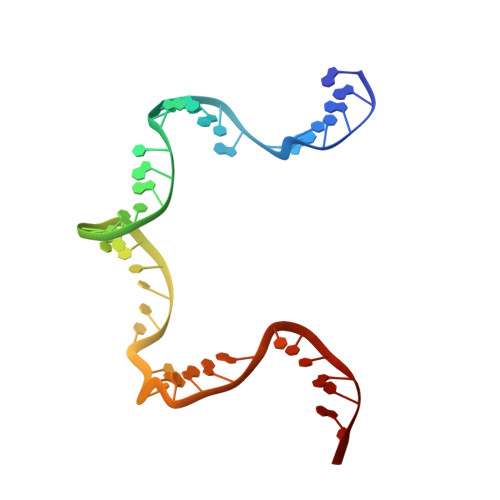
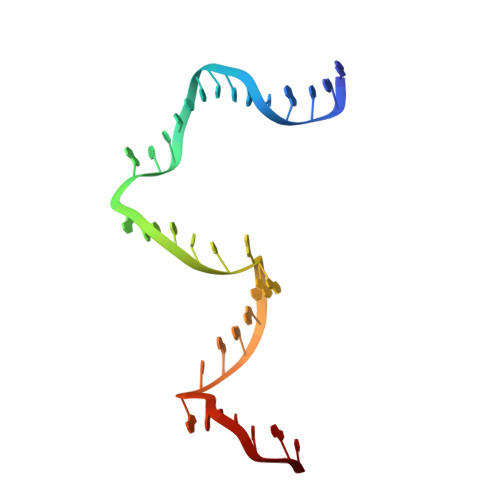
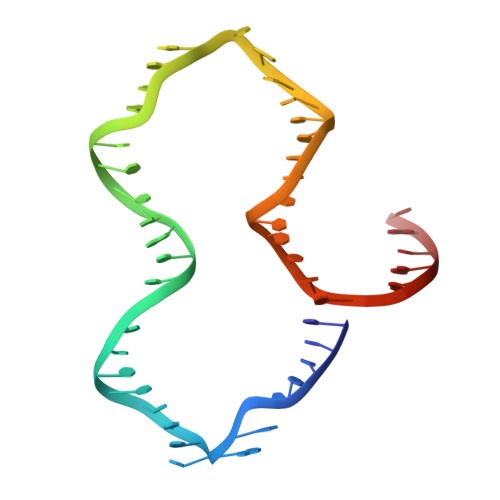
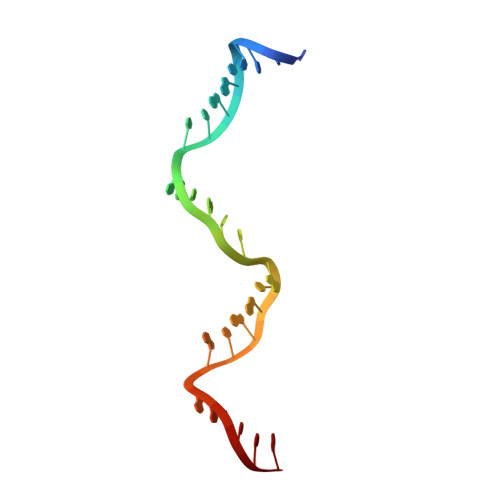
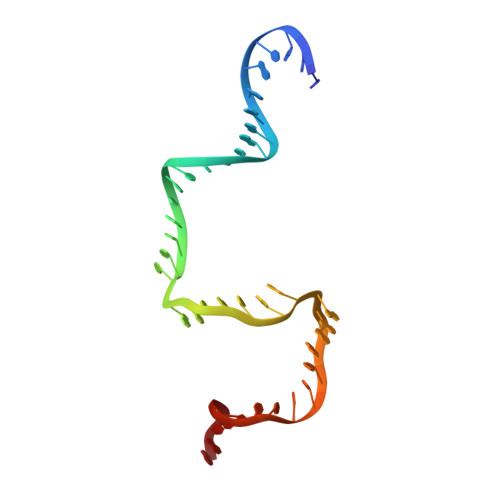
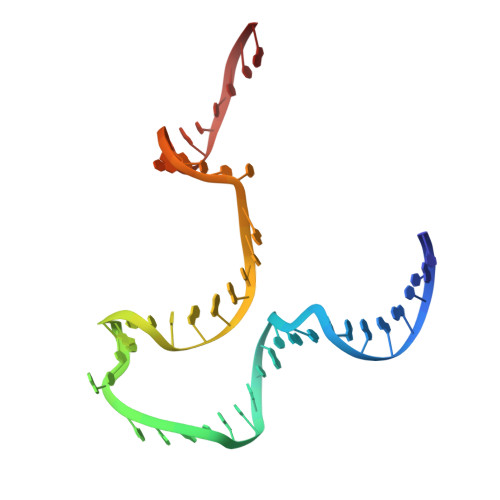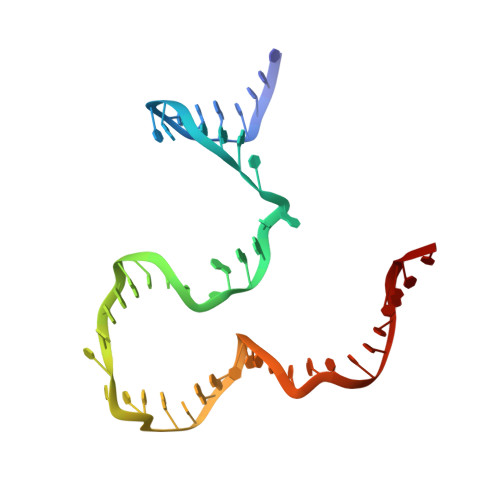Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Kube, M., Kohler, F., Feigl, E., Nagel-Yuksel, B., Willner, E.M., Funke, J.J., Gerling, T., Stommer, P., Honemann, M.N., Martin, T.G., Scheres, S.H.W., Dietz, H.(2020) Nat Commun 11: 6229-6229
- PubMed: 33277481
- DOI: https://doi.org/10.1038/s41467-020-20020-7
- Primary Citation of Related Structures:
7ARE, 7ARQ, 7ART, 7ARV, 7ARY, 7AS5 - PubMed Abstract:
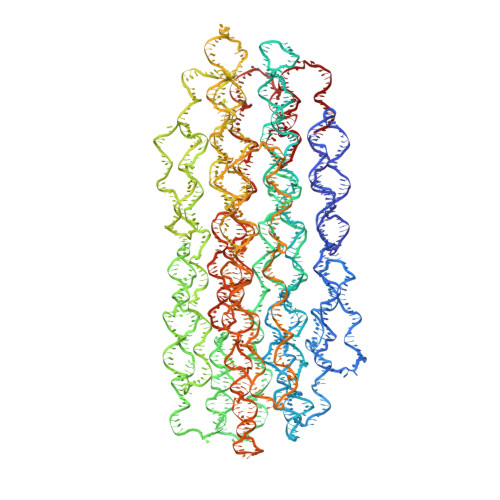
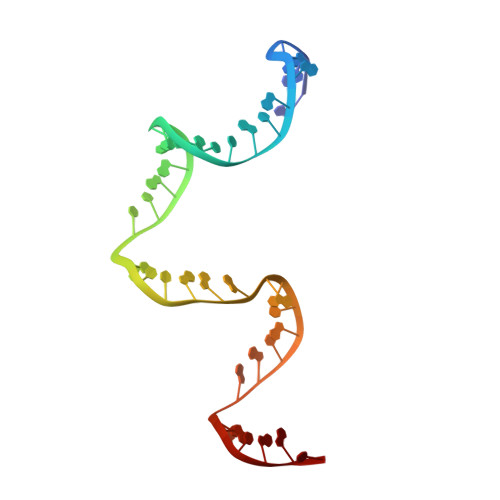
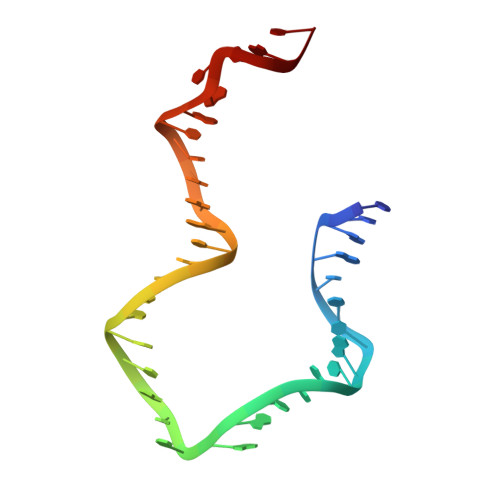
The methods of DNA nanotechnology enable the rational design of custom shapes that self-assemble in solution from sets of DNA molecules. DNA origami, in which a long template DNA single strand is folded by many short DNA oligonucleotides, can be employed to make objects comprising hundreds of unique DNA strands and thousands of base pairs, thus in principle providing many degrees of freedom for modelling complex objects of defined 3D shapes and sizes. Here, we address the problem of accurate structural validation of DNA objects in solution with cryo-EM based methodologies. By taking into account structural fluctuations, we can determine structures with improved detail compared to previous work. To interpret the experimental cryo-EM maps, we present molecular-dynamics-based methods for building pseudo-atomic models in a semi-automated fashion. Among other features, our data allows discerning details such as helical grooves, single-strand versus double-strand crossovers, backbone phosphate positions, and single-strand breaks. Obtaining this higher level of detail is a step forward that now allows designers to inspect and refine their designs with base-pair level interventions.
Organizational Affiliation:
Physik Department, Technische Universität München, Garching, Germany.