Fucosylated antimicrobial peptide SB6 in complex with the lectin LecRSL from Ralstonia solanacearum at 2.4 Angstrom resolution
Baeriswyl, S., Stocker, A., Reymond, J.-L.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fucose-binding lectin protein | 91 | Ralstonia solanacearum | Mutation(s): 0 Gene Names: E7Z57_08365, RSP795_21825, RSP822_19650, RUN39_v1_50103 | 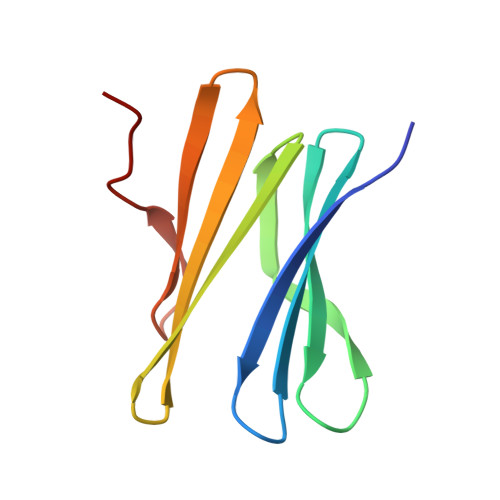 | |
UniProt | |||||
Find proteins for A0A0S4TLR1 (Ralstonia solanacearum) Explore A0A0S4TLR1 Go to UniProtKB: A0A0S4TLR1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0S4TLR1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| SB6 | C [auth I], F [auth K], N [auth R] | 12 | synthetic construct | Mutation(s): 0 | 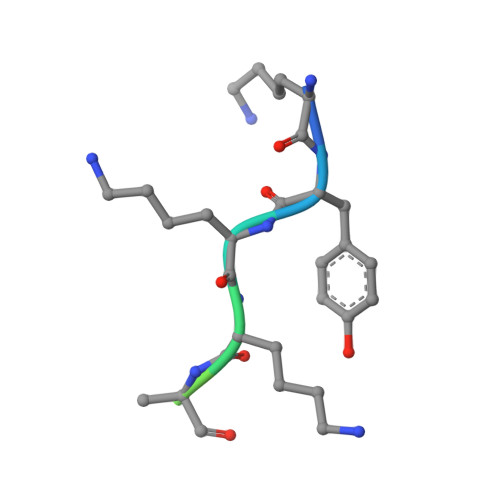 |
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| SB6 | D [auth J], G [auth L], I [auth M], K [auth O], M [auth Q] | 13 | synthetic construct | Mutation(s): 0 | 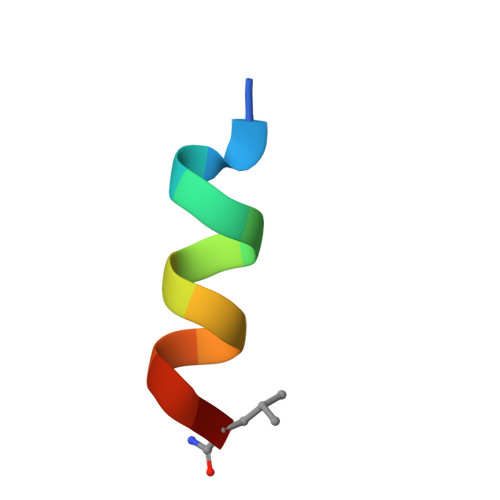 |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ZDC (Subject of Investigation/LOI) Query on ZDC | AA [auth R] O [auth A] Q [auth A] R [auth I] S [auth J] | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid C8 H14 O6 YTZUDUWVQZSKNN-OASCRQMUSA-N |  | ||
| DLY (Subject of Investigation/LOI) Query on DLY | P [auth A] | D-LYSINE C6 H14 N2 O2 KDXKERNSBIXSRK-RXMQYKEDSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 100.51 | α = 90 |
| b = 100.51 | β = 90 |
| c = 271.653 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Swiss National Science Foundation | Switzerland | -- |