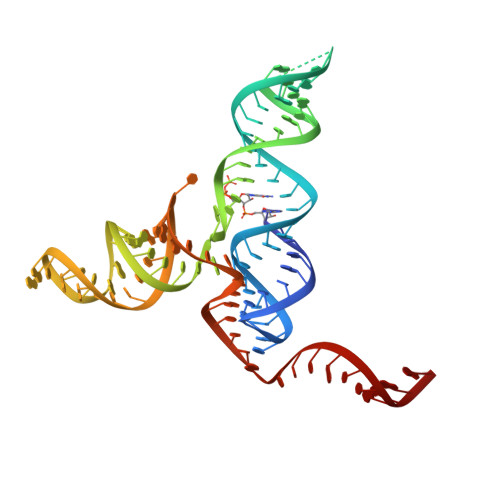
Two riboswitch classes that share a common ligand-binding fold show major differences in the ability to accommodate mutations.
Srivastava, Y., Akinyemi, O., Rohe, T.C., Pritchett, E.M., Baker, C.D., Sharma, A., Jenkins, J.L., Mathews, D.H., Wedekind, J.E.(2024) Nucleic Acids Res 52: 13152-13173
- PubMed: 39413212
- DOI: https://doi.org/10.1093/nar/gkae886
- Primary Citation of Related Structures:
6XKN, 6XKO, 8TOZ, 8VPV - PubMed Abstract:
Riboswitches are structured RNAs that sense small molecules to control expression. Prequeuosine1 (preQ1)-sensing riboswitches comprise three classes (I, II and III) that adopt distinct folds. Despite this difference, class II and III riboswitches each use 10 identical nucleotides to bind the preQ1 metabolite. Previous class II studies showed high sensitivity to binding-pocket mutations, which reduced preQ1 affinity and impaired function. Here, we introduced four equivalent mutations into a class III riboswitch, which maintained remarkably tight preQ1 binding. Co-crystal structures of each class III mutant showed compensatory interactions that preserve the fold. Chemical modification analysis revealed localized RNA flexibility changes for each mutant, but molecular dynamics (MD) simulations suggested that each mutation was not overtly destabilizing. Although impaired, class III mutants retained tangible gene-regulatory activity in bacteria compared to equivalent preQ1-II variants; mutations in the preQ1-pocket floor were tolerated better than wall mutations. Principal component analysis of MD trajectories suggested that the most functionally deleterious wall mutation samples different motions compared to wildtype. Overall, the results reveal that formation of compensatory interactions depends on the context of mutations within the overall fold and that functionally deleterious mutations can alter long-range correlated motions that link the riboswitch binding pocket with distal gene-regulatory sequences.
Organizational Affiliation:
Department of Biochemistry and Biophysics, University of Rochester School of Medicine and Dentistry, 601 Elmwood Ave MC 712, Rochester, NY 14642, USA.