New insights into Class III PreQ1 metabolite binding
Srivastava, Y., Jermaine, J.L., Wedekind, J.E.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| RNA (101-MER) | 100 | Faecalibacterium prausnitzii | 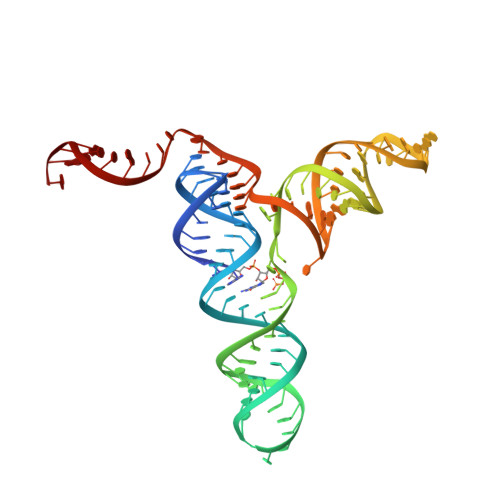 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PRF (Subject of Investigation/LOI) Query on PRF | B [auth A] | 7-DEAZA-7-AMINOMETHYL-GUANINE C7 H9 N5 O MEYMBLGOKYDGLZ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 82.685 | α = 90 |
| b = 82.685 | β = 90 |
| c = 282.007 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM63162-14 |