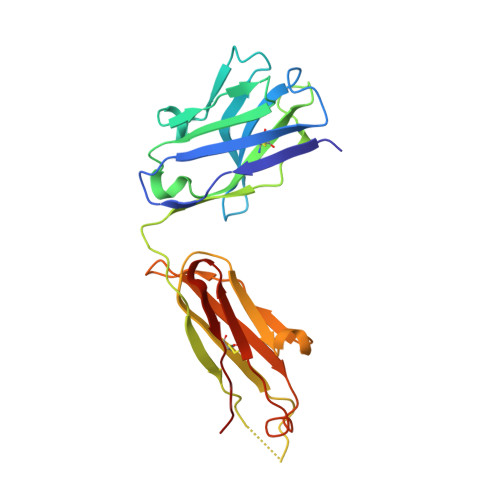
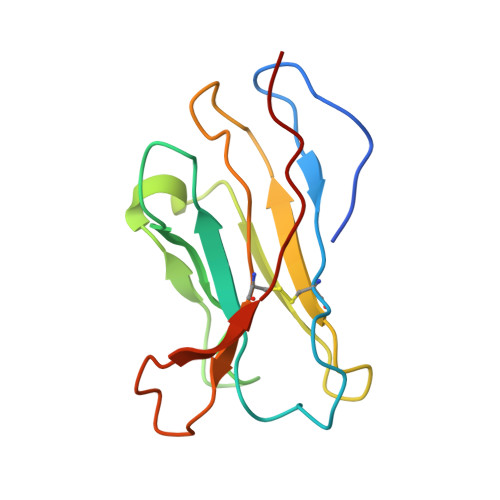
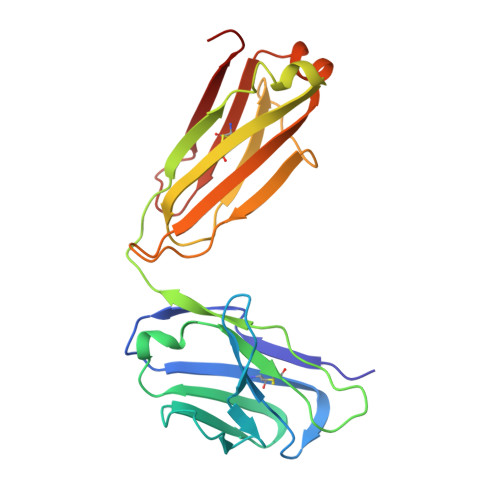
A GPC2 antibody-drug conjugate is efficacious against neuroblastoma and small-cell lung cancer via binding a conformational epitope.
Raman, S., Buongervino, S.N., Lane, M.V., Zhelev, D.V., Zhu, Z., Cui, H., Martinez, B., Martinez, D., Wang, Y., Upton, K., Patel, K., Rathi, K.S., Navia, C.T., Harmon, D.B., Li, Y., Pawel, B., Dimitrov, D.S., Maris, J.M., Julien, J.P., Bosse, K.R.(2021) Cell Rep Med 2: 100344-100344
- PubMed: 34337560
- DOI: https://doi.org/10.1016/j.xcrm.2021.100344
- Primary Citation of Related Structures:
6WJL - PubMed Abstract:
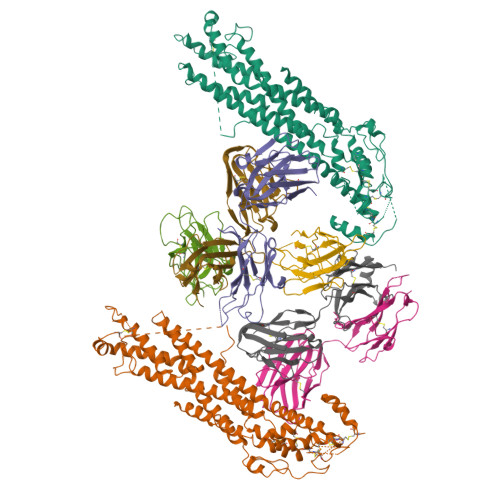
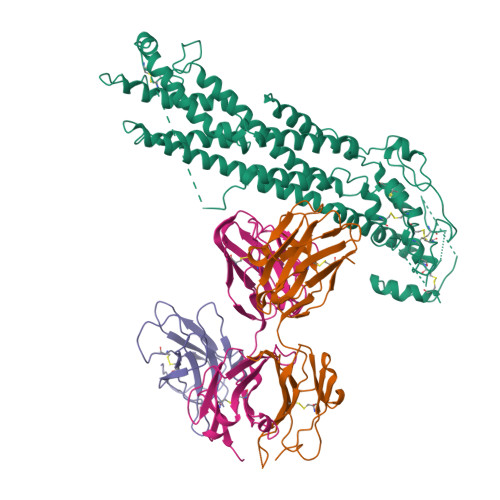
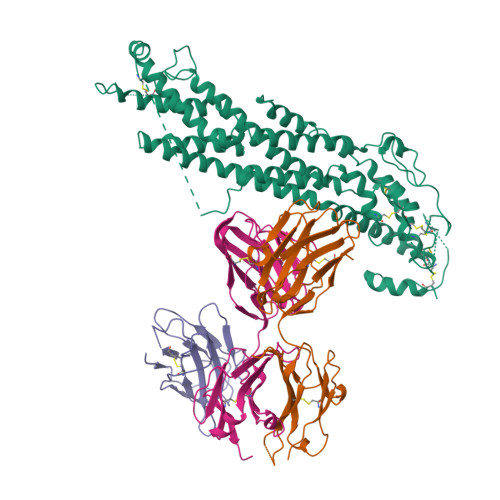
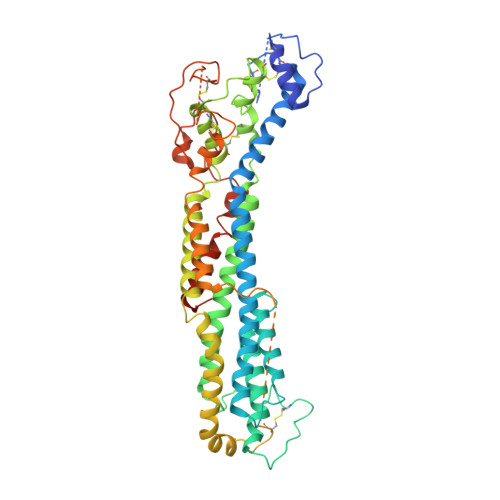
Glypican 2 (GPC2) is a MYCN-regulated, differentially expressed cell-surface oncoprotein and target for immune-based therapies in neuroblastoma. Here, we build on GPC2's immunotherapeutic attributes by finding that it is also a highly expressed, MYCN-driven oncoprotein on small-cell lung cancers (SCLCs), with significantly enriched expression in both the SCLC and neuroblastoma stem cell compartment.By solving the crystal structure of the D3-GPC2-Fab/GPC2 complex at 3.3 Å resolution, we further illustrate that the GPC2-directed antibody-drug conjugate (ADC; D3-GPC2-PBD), that links a human GPC2 antibody (D3) to DNA-damaging pyrrolobenzodiazepine (PBD) dimers, binds a tumor-specific, conformation-dependent epitope of the core GPC2 extracellular domain. We then show that this ADC induces durable neuroblastoma and SCLC tumor regression via induction of DNA damage, apoptosis, and bystander cell killing, notably with no signs of ADC-induced in vivo toxicity. These studies provide preclinical data to support the clinical translation of ADCs targeting GPC2.
Organizational Affiliation:
Program in Molecular Medicine, Hospital for Sick Children Research Institute, Toronto, ON M5G 0A4, Canada.