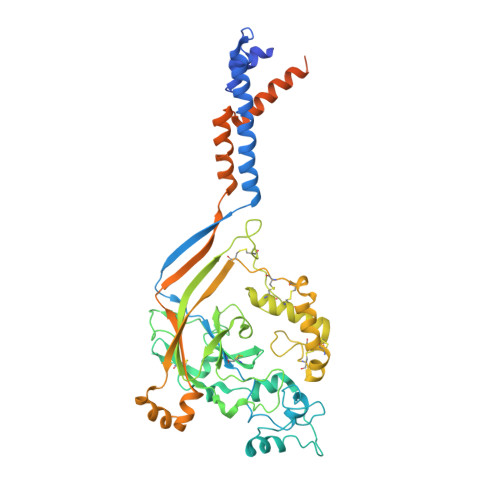
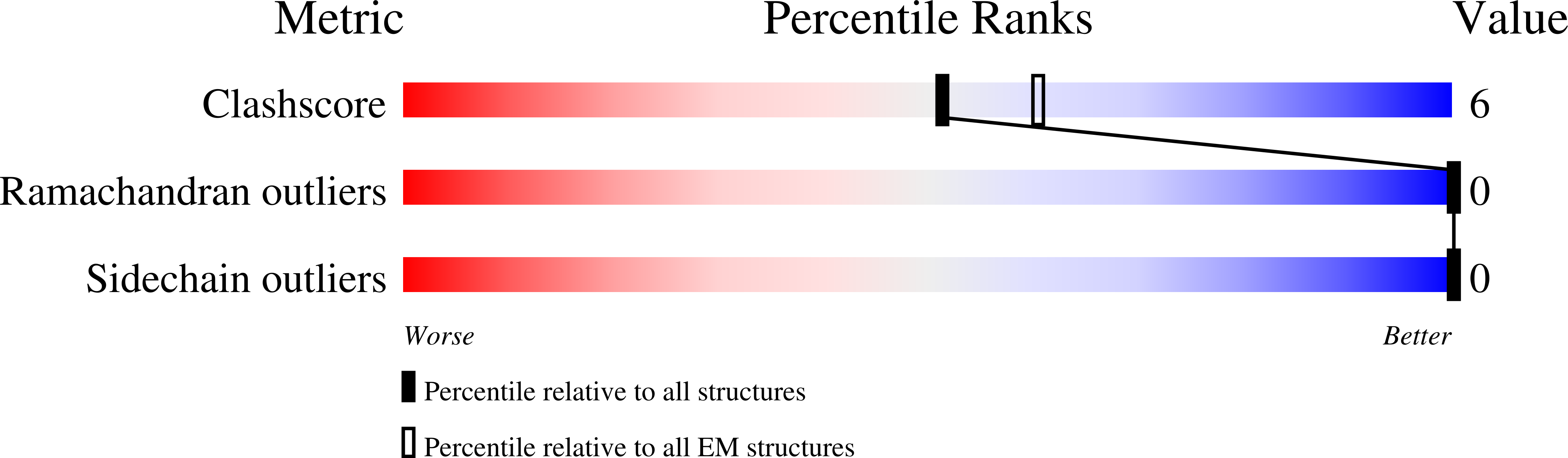The His-Gly motif of acid-sensing ion channels resides in a reentrant 'loop' implicated in gating and ion selectivity.
Yoder, N., Gouaux, E.(2020) Elife 9
- PubMed: 32496192
- DOI: https://doi.org/10.7554/eLife.56527
- Primary Citation of Related Structures:
6VTK, 6VTL - PubMed Abstract:
Acid-sensing ion channels (ASICs) are proton-gated members of the epithelial sodium channel/degenerin (ENaC/DEG) superfamily of ion channels and are expressed throughout the central and peripheral nervous systems. The homotrimeric splice variant ASIC1a has been implicated in nociception, fear memory, mood disorders and ischemia. Here, we extract full-length chicken ASIC1 (cASIC1) from cell membranes using styrene maleic acid (SMA) copolymer, elucidating structures of ASIC1 channels in both high pH resting and low pH desensitized conformations by single-particle cryo-electron microscopy (cryo-EM). The structures of resting and desensitized channels reveal a reentrant loop at the amino terminus of ASIC1 that includes the highly conserved 'His-Gly' (HG) motif. The reentrant loop lines the lower ion permeation pathway and buttresses the 'Gly-Ala-Ser' (GAS) constriction, thus providing a structural explanation for the role of the His-Gly dipeptide in the structure and function of ASICs.
Organizational Affiliation:
Vollum Institute, Oregon Health & Science University, Portland, United States.