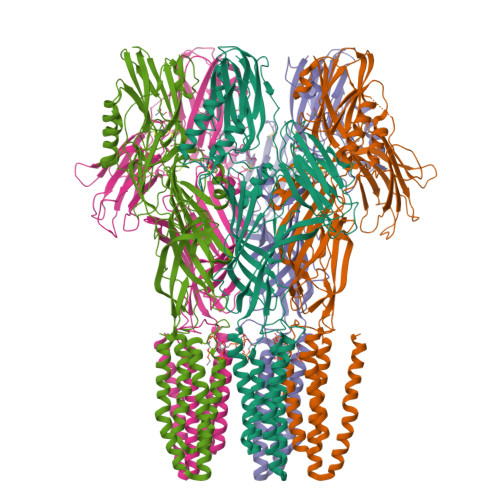
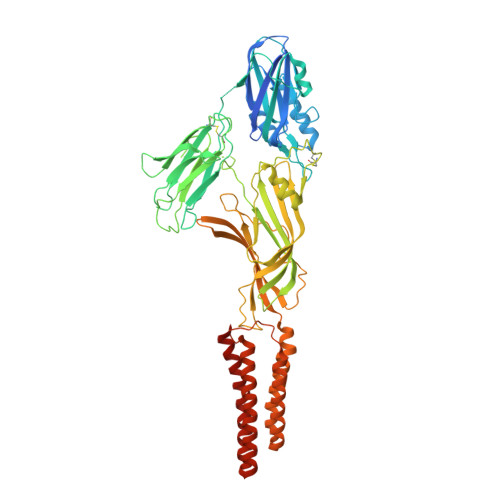
Structural basis for allosteric transitions of a multidomain pentameric ligand-gated ion channel.
Hu, H., Howard, R.J., Bastolla, U., Lindahl, E., Delarue, M.(2020) Proc Natl Acad Sci U S A 117: 13437-13446
- PubMed: 32482881
- DOI: https://doi.org/10.1073/pnas.1922701117
- Primary Citation of Related Structures:
6V4A, 6V4B, 6V4S - PubMed Abstract:
Pentameric ligand-gated ion channels (pLGICs) are allosteric receptors that mediate rapid electrochemical signal transduction in the animal nervous system through the opening of an ion pore upon binding of neurotransmitters. Orthologs have been found and characterized in prokaryotes and they display highly similar structure-function relationships to eukaryotic pLGICs; however, they often encode greater architectural diversity involving additional amino-terminal domains (NTDs). Here we report structural, functional, and normal-mode analysis of two conformational states of a multidomain pLGIC, called DeCLIC, from a Desulfofustis deltaproteobacterium, including a periplasmic NTD fused to the conventional ligand-binding domain (LBD). X-ray structure determination revealed an NTD consisting of two jelly-roll domains interacting across each subunit interface. Binding of Ca 2+ at the LBD subunit interface was associated with a closed transmembrane pore, with resolved monovalent cations intracellular to the hydrophobic gate. Accordingly, DeCLIC-injected oocytes conducted currents only upon depletion of extracellular Ca 2+ ; these were insensitive to quaternary ammonium block. Furthermore, DeCLIC crystallized in the absence of Ca 2+ with a wide-open pore and remodeled periplasmic domains, including increased contacts between the NTD and classic LBD agonist-binding sites. Functional, structural, and dynamical properties of DeCLIC paralleled those of sTeLIC, a pLGIC from another symbiotic prokaryote. Based on these DeCLIC structures, we would reclassify the previous structure of bacterial ELIC (the first high-resolution structure of a pLGIC) as a "locally closed" conformation. Taken together, structures of DeCLIC in multiple conformations illustrate dramatic conformational state transitions and diverse regulatory mechanisms available to ion channels in pLGICs, particularly involving Ca 2+ modulation and periplasmic NTDs.
Organizational Affiliation:
Unité Dynamique Structurale des Macromolécules, Institut Pasteur, UMR 3528, CNRS, 75015 Paris, France.