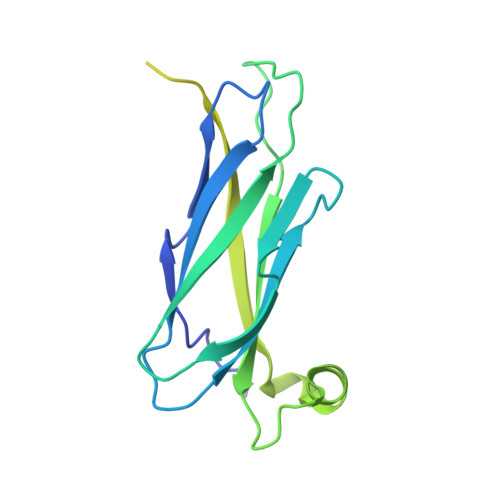
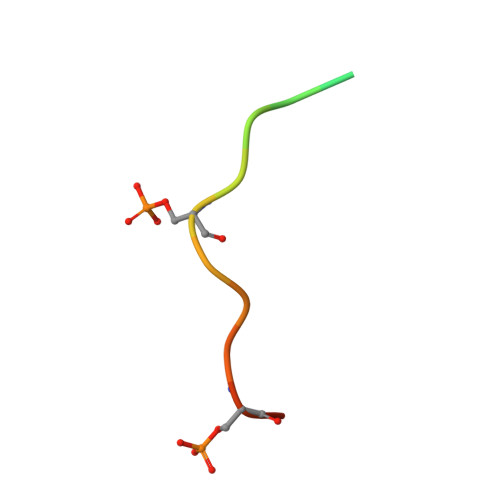
FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Di Mattia, T., Martinet, A., Ikhlef, S., McEwen, A.G., Nomine, Y., Wendling, C., Poussin-Courmontagne, P., Voilquin, L., Eberling, P., Ruffenach, F., Cavarelli, J., Slee, J., Levine, T.P., Drin, G., Tomasetto, C., Alpy, F.(2020) EMBO J 39: e104369-e104369
- PubMed: 33124732
- DOI: https://doi.org/10.15252/embj.2019104369
- Primary Citation of Related Structures:
6TQR, 6TQS, 6TQT, 6TQU - PubMed Abstract:
Organelles are physically connected in membrane contact sites. The endoplasmic reticulum possesses three major receptors, VAP-A, VAP-B, and MOSPD2, which interact with proteins at the surface of other organelles to build contacts. VAP-A, VAP-B, and MOSPD2 contain an MSP domain, which binds a motif named FFAT (two phenylalanines in an acidic tract). In this study, we identified a non-conventional FFAT motif where a conserved acidic residue is replaced by a serine/threonine. We show that phosphorylation of this serine/threonine is critical for non-conventional FFAT motifs (named Phospho-FFAT) to be recognized by the MSP domain. Moreover, structural analyses of the MSP domain alone or in complex with conventional and Phospho-FFAT peptides revealed new mechanisms of interaction. Based on these new insights, we produced a novel prediction algorithm, which expands the repertoire of candidate proteins with a Phospho-FFAT that are able to create membrane contact sites. Using a prototypical tethering complex made by STARD3 and VAP, we showed that phosphorylation is instrumental for the formation of ER-endosome contacts, and their sterol transfer function. This study reveals that phosphorylation acts as a general switch for inter-organelle contacts.
Organizational Affiliation:
Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), Illkirch, France.