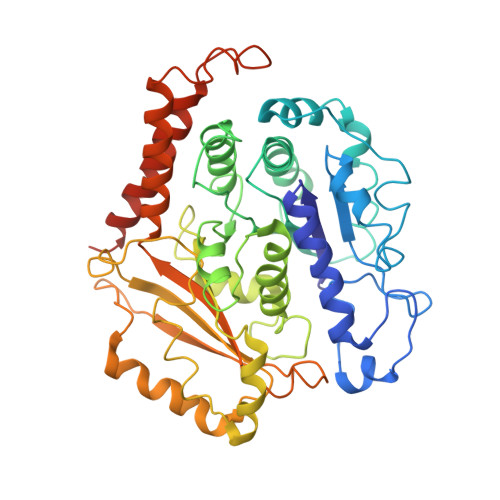
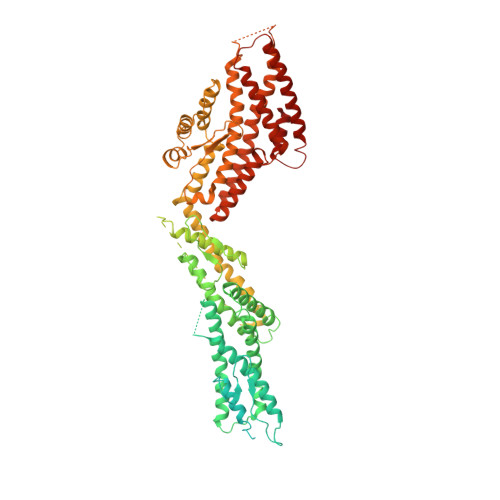
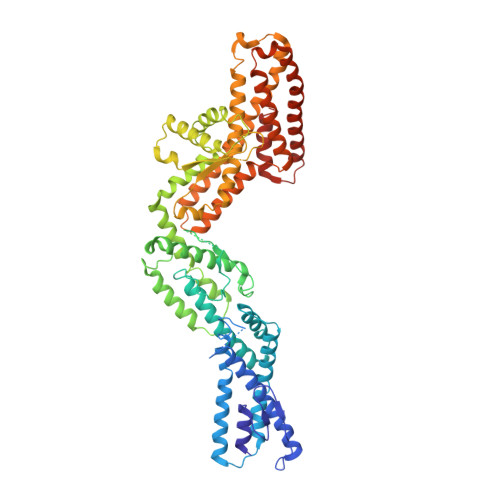
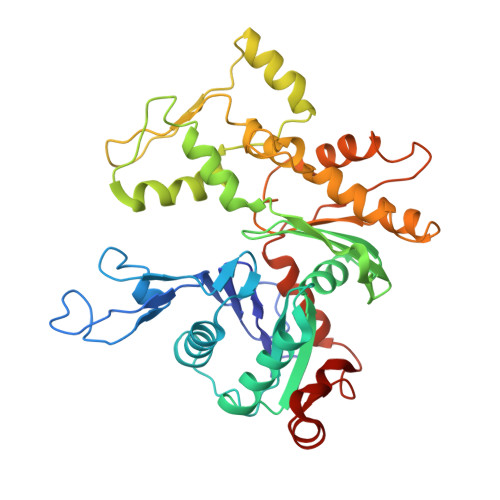
Insights into the assembly and activation of the microtubule nucleator gamma-TuRC.
Liu, P., Zupa, E., Neuner, A., Bohler, A., Loerke, J., Flemming, D., Ruppert, T., Rudack, T., Peter, C., Spahn, C., Gruss, O.J., Pfeffer, S., Schiebel, E.(2020) Nature 578: 467-471
- PubMed: 31856152
- DOI: https://doi.org/10.1038/s41586-019-1896-6
- Primary Citation of Related Structures:
6TF9 - PubMed Abstract:
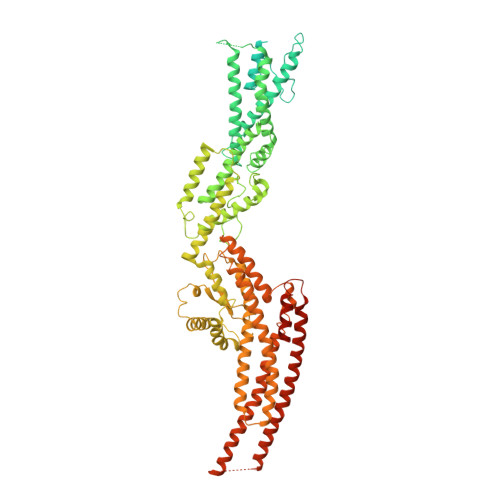
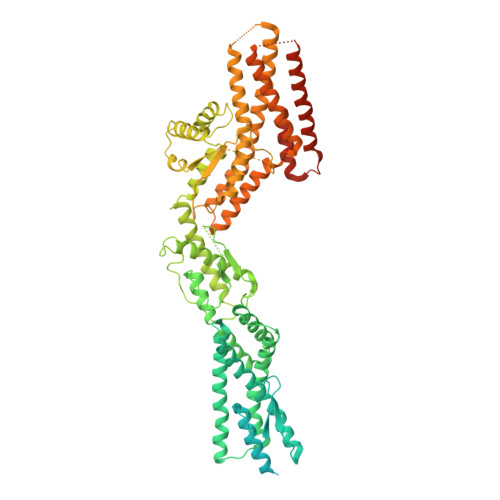
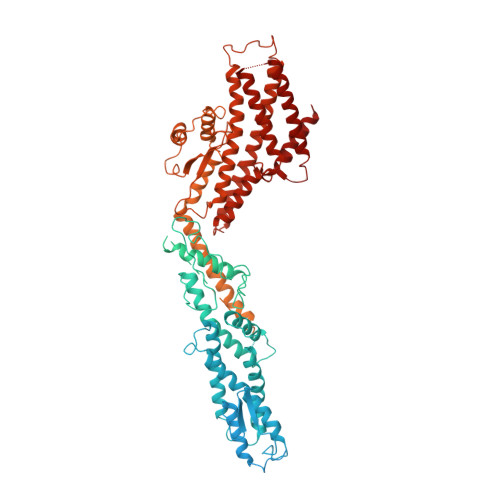
Microtubules are dynamic polymers of α- and β-tubulin and have crucial roles in cell signalling, cell migration, intracellular transport and chromosome segregation 1 . They assemble de novo from αβ-tubulin dimers in an essential process termed microtubule nucleation. Complexes that contain the protein γ-tubulin serve as structural templates for the microtubule nucleation reaction 2 . In vertebrates, microtubules are nucleated by the 2.2-megadalton γ-tubulin ring complex (γ-TuRC), which comprises γ-tubulin, five related γ-tubulin complex proteins (GCP2-GCP6) and additional factors 3 . GCP6 is unique among the GCP proteins because it carries an extended insertion domain of unknown function. Our understanding of microtubule formation in cells and tissues is limited by a lack of high-resolution structural information on the γ-TuRC. Here we present the cryo-electron microscopy structure of γ-TuRC from Xenopus laevis at 4.8 Å global resolution, and identify a 14-spoked arrangement of GCP proteins and γ-tubulins in a partially flexible open left-handed spiral with a uniform sequence of GCP variants. By forming specific interactions with other GCP proteins, the GCP6-specific insertion domain acts as a scaffold for the assembly of the γ-TuRC. Unexpectedly, we identify actin as a bona fide structural component of the γ-TuRC with functional relevance in microtubule nucleation. The spiral geometry of γ-TuRC is suboptimal for microtubule nucleation and a controlled conformational rearrangement of the γ-TuRC is required for its activation. Collectively, our cryo-electron microscopy reconstructions provide detailed insights into the molecular organization, assembly and activation mechanism of vertebrate γ-TuRC, and will serve as a framework for the mechanistic understanding of fundamental biological processes associated with microtubule nucleation, such as meiotic and mitotic spindle formation and centriole biogenesis 4 .
Organizational Affiliation:
Zentrum für Molekulare Biologie, Universität Heidelberg, DKFZ-ZMBH Allianz, Heidelberg, Germany.