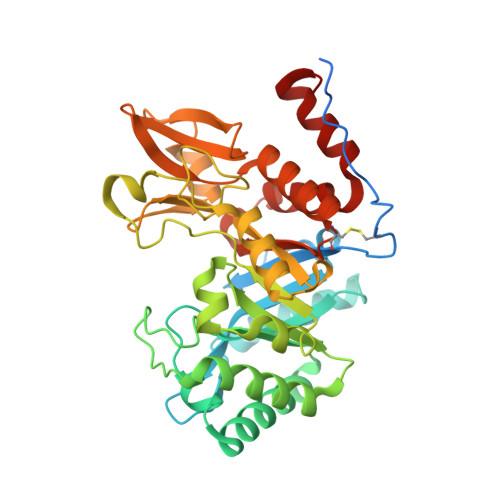
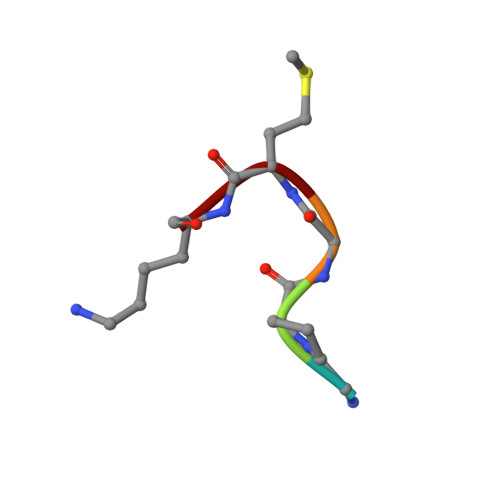
Crystal structures of the GH18 domain of the bifunctional peroxiredoxin-chitinase CotE from Clostridium difficile.
Whittingham, J.L., Hanai, S., Brannigan, J.A., Ferreira, W.T., Dodson, E.J., Turkenburg, J.P., Cartwright, J., Cutting, S.M., Wilkinson, A.J.(2020) Acta Crystallogr F Struct Biol Commun 76: 241-249
- PubMed: 32510464
- DOI: https://doi.org/10.1107/S2053230X20006147
- Primary Citation of Related Structures:
6T9M, 6TSB - PubMed Abstract:
CotE is a coat protein that is present in the spores of Clostridium difficile, an obligate anaerobic bacterium and a pathogen that is a leading cause of antibiotic-associated diarrhoea in hospital patients. Spores serve as the agents of disease transmission, and CotE has been implicated in their attachment to the gut epithelium and subsequent colonization of the host. CotE consists of an N-terminal peroxiredoxin domain and a C-terminal chitinase domain. Here, a C-terminal fragment of CotE comprising residues 349-712 has been crystallized and its structure has been determined to reveal a core eight-stranded β-barrel fold with a neighbouring subdomain containing a five-stranded β-sheet. A prominent groove running across the top of the barrel is lined by residues that are conserved in family 18 glycosyl hydrolases and which participate in catalysis. Electron density identified in the groove defines the pentapeptide Gly-Pro-Ala-Met-Lys derived from the N-terminus of the protein following proteolytic cleavage to remove an affinity-purification tag. These observations suggest the possibility of designing peptidomimetics to block C. difficile transmission.
Organizational Affiliation:
Structural Biology Laboratory, York Biomedical Research Institute, Department of Chemistry, University of York, York YO10 5DD, United Kingdom.