Aequorea's secrets revealed: New fluorescent proteins with unique properties for bioimaging and biosensing.
Lambert, G.G., Depernet, H., Gotthard, G., Schultz, D.T., Navizet, I., Lambert, T., Adams, S.R., Torreblanca-Zanca, A., Chu, M., Bindels, D.S., Levesque, V., Nero Moffatt, J., Salih, A., Royant, A., Shaner, N.C.(2020) PLoS Biol 18: e3000936-e3000936
- PubMed: 33137097
- DOI: https://doi.org/10.1371/journal.pbio.3000936
- Primary Citation of Related Structures:
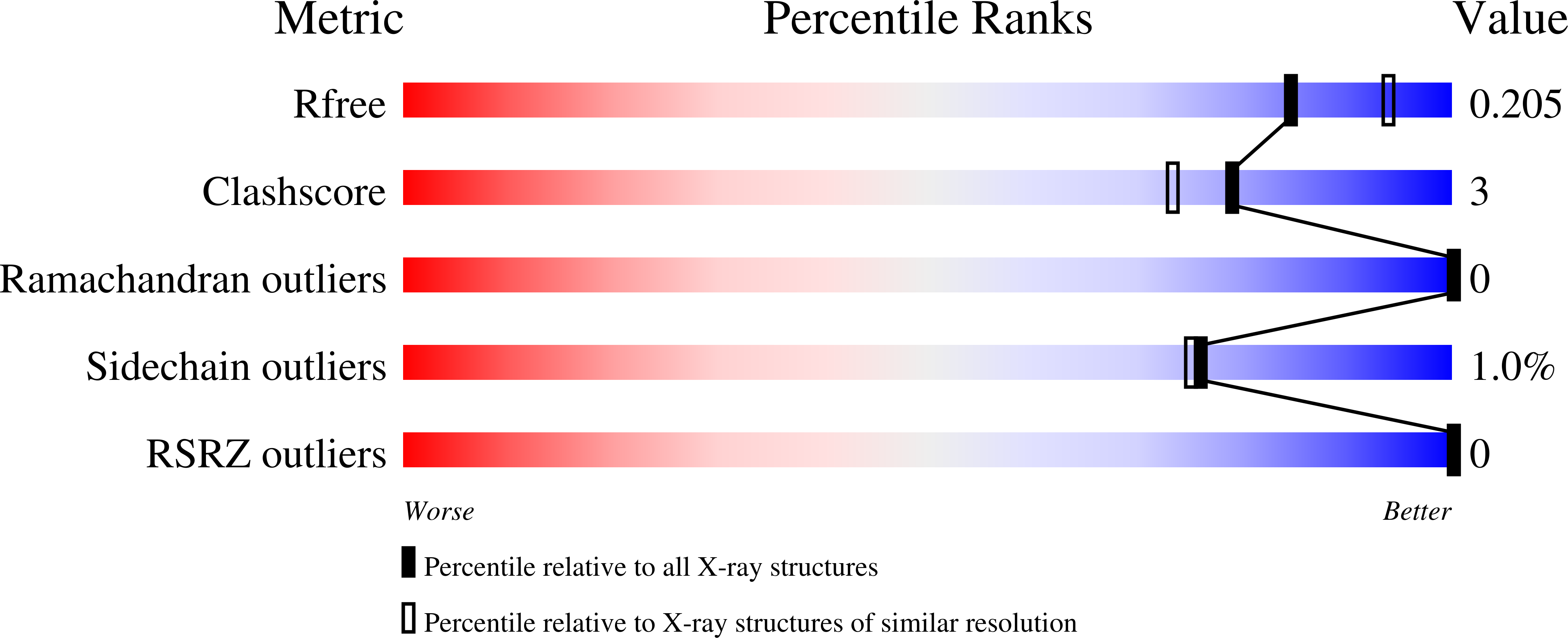
6S67, 6S68 - PubMed Abstract:
Using mRNA sequencing and de novo transcriptome assembly, we identified, cloned, and characterized 9 previously undiscovered fluorescent protein (FP) homologs from Aequorea victoria and a related Aequorea species, with most sequences highly divergent from A. victoria green fluorescent protein (avGFP). Among these FPs are the brightest green fluorescent protein (GFP) homolog yet characterized and a reversibly photochromic FP that responds to UV and blue light. Beyond green emitters, Aequorea species express purple- and blue-pigmented chromoproteins (CPs) with absorbances ranging from green to far-red, including 2 that are photoconvertible. X-ray crystallography revealed that Aequorea CPs contain a chemically novel chromophore with an unexpected crosslink to the main polypeptide chain. Because of the unique attributes of several of these newly discovered FPs, we expect that Aequorea will, once again, give rise to an entirely new generation of useful probes for bioimaging and biosensing.
Organizational Affiliation:
Department of Neurosciences, Center for Research in Biological Systems, University of California San Diego School of Medicine, La Jolla, California, United States of America.