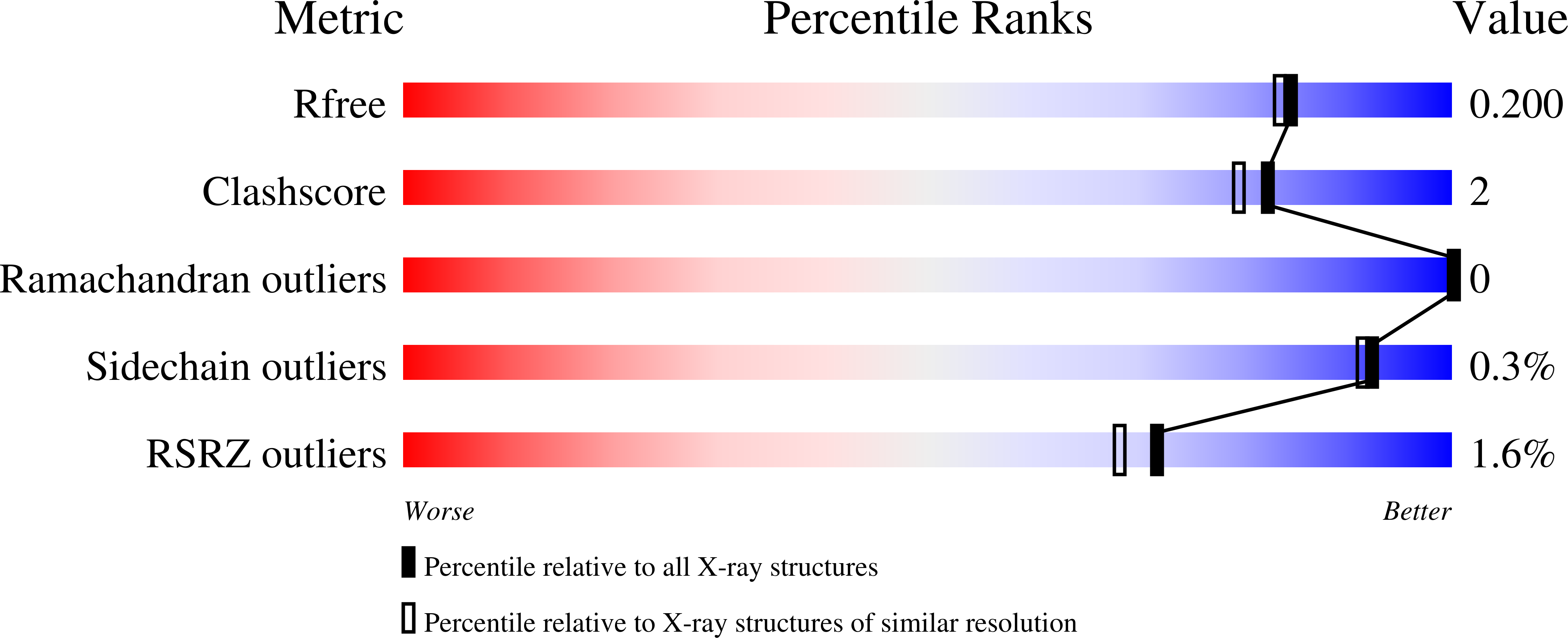
The minimal structure for iodotyrosine deiodinase function is defined by an outlier protein from the thermophilic bacterium Thermotoga neapolitana.
Sun, Z., Xu, B., Spisak, S., Kavran, J.M., Rokita, S.E.(2021) J Biol Chem 297: 101385-101385
- PubMed: 34748729
- DOI: https://doi.org/10.1016/j.jbc.2021.101385
- Primary Citation of Related Structures:
6PZ0, 6Q1L - PubMed Abstract:
The nitroreductase superfamily of enzymes encompasses many flavin mononucleotide (FMN)-dependent catalysts promoting a wide range of reactions. All share a common core consisting of an FMN-binding domain, and individual subgroups additionally contain one to three sequence extensions radiating from defined positions within this core to support their unique catalytic properties. To identify the minimum structure required for activity in the iodotyrosine deiodinase subgroup of this superfamily, attention was directed to a representative from the thermophilic organism Thermotoga neapolitana (TnIYD). This representative was selected based on its status as an outlier of the subgroup arising from its deficiency in certain standard motifs evident in all homologues from mesophiles. We found that TnIYD lacked a typical N-terminal sequence and one of its two characteristic sequence extensions, neither of which was found to be necessary for activity. We also show that TnIYD efficiently promotes dehalogenation of iodo-, bromo-, and chlorotyrosine, analogous to related deiodinases (IYDs) from humans and other mesophiles. In addition, 2-iodophenol is a weak substrate for TnIYD as it was for all other IYDs characterized to date. Consistent with enzymes from thermophilic organisms, we observed that TnIYD adopts a compact fold and low surface area compared with IYDs from mesophilic organisms. The insights gained from our investigations on TnIYD demonstrate the advantages of focusing on sequences that diverge from conventional standards to uncover the minimum essentials for activity. We conclude that TnIYD now represents a superior starting structure for future efforts to engineer a stable dehalogenase targeting halophenols of environmental concern.
Organizational Affiliation:
Department of Chemistry, Johns Hopkins University, Baltimore, Maryland, USA.