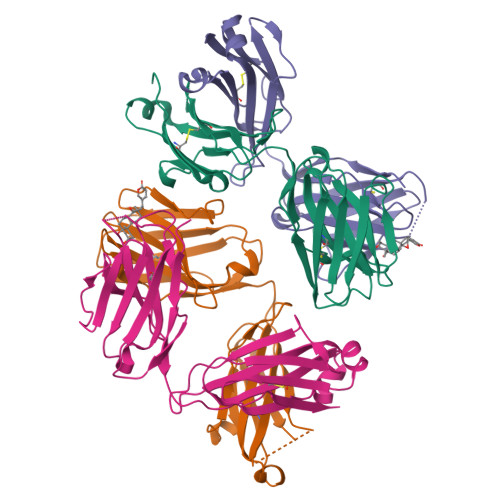
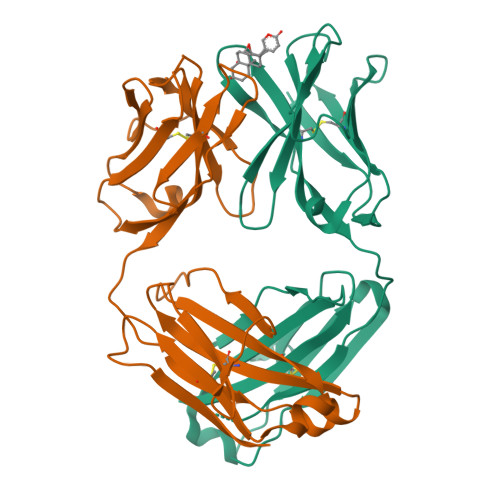
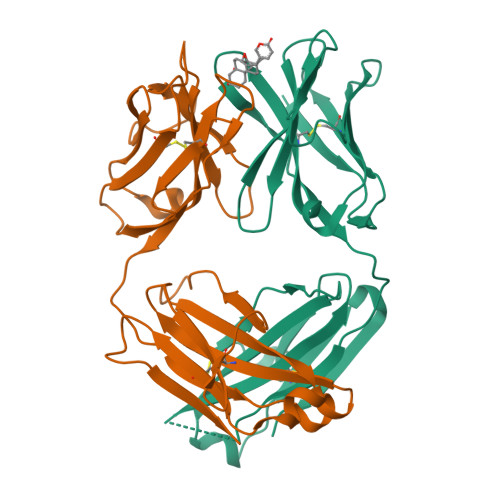
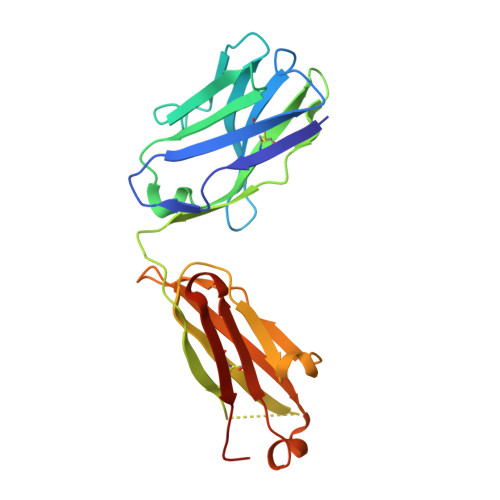
Kappa-on-Heavy (KoH) bodies are a distinct class of fully-human antibody-like therapeutic agents with antigen-binding properties.
Macdonald, L.E., Meagher, K.A., Franklin, M.C., Levenkova, N., Hansen, J., Badithe, A.T., Zhong, M., Krueger, P., Rafique, A., Tu, N., Shevchuk, J., Wadhwa, S., Ehrlich, G., Bautista, J., Grant, C., Esau, L., Poueymirou, W.T., Auerbach, W., Morton, L., Babb, R., Chen, G., Huang, T., MacDonald, D., Graham, K., Gurer, C., Voronina, V.A., McWhirter, J.R., Guo, C., Yancopoulos, G.D., Murphy, A.J.(2020) Proc Natl Acad Sci U S A 117: 292-299
- PubMed: 31879340
- DOI: https://doi.org/10.1073/pnas.1901734117
- Primary Citation of Related Structures:
6PYC, 6PYD - PubMed Abstract:
We describe a Kappa-on-Heavy (KoH) mouse that produces a class of highly diverse, fully human, antibody-like agents. This mouse was made by replacing the germline variable sequences of both the Ig heavy-chain (IgH) and Ig kappa (IgK) loci with the human IgK germline variable sequences, producing antibody-like molecules with an antigen binding site made up of 2 kappa variable domains. These molecules, named KoH bodies, structurally mimic naturally existing Bence-Jones light-chain dimers in their variable domains and remain wild-type in their antibody constant domains. Unlike artificially diversified, nonimmunoglobulin alternative scaffolds (e.g., DARPins), KoH bodies consist of a configuration of normal Ig scaffolds that undergo natural diversification in B cells. Monoclonal KoH bodies have properties similar to those of conventional antibodies but exhibit an enhanced ability to bind small molecules such as the endogenous cardiotonic steroid marinobufagenin (MBG) and nicotine. A comparison of crystal structures of MBG bound to a KoH Fab versus a conventional Fab showed that the KoH body has a much deeper binding pocket, allowing MBG to be held 4 Å further down into the combining site between the 2 variable domains.
Organizational Affiliation:
Regeneron Tech Centers, Regeneron Pharmaceuticals, Inc., Tarrytown, NY 10591 lynn.macdonald@regeneron.com.