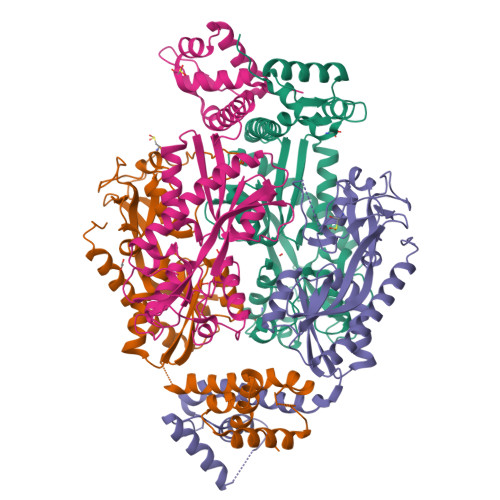
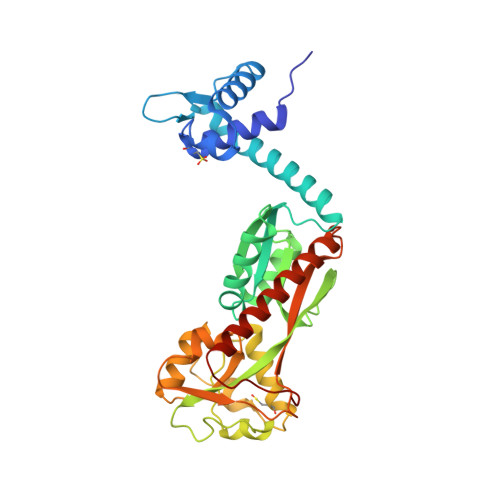
Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2sensing.
Pedre, B., Young, D., Charlier, D., Mourenza, A., Rosado, L.A., Marcos-Pascual, L., Wahni, K., Martens, E., G de la Rubia, A., Belousov, V.V., Mateos, L.M., Messens, J.(2018) Proc Natl Acad Sci U S A 115: E11623-E11632
- PubMed: 30463959
- DOI: https://doi.org/10.1073/pnas.1807954115
- Primary Citation of Related Structures:
6G1B, 6G1D, 6G4R - PubMed Abstract:
Hydrogen peroxide (H 2 O 2 ) is a strong oxidant capable of oxidizing cysteinyl thiolates, yet only a few cysteine-containing proteins have exceptional reactivity toward H 2 O 2 One such example is the prokaryotic transcription factor OxyR, which controls the antioxidant response in bacteria, and which specifically and rapidly reduces H 2 O 2 In this study, we present crystallographic evidence for the H 2 O 2 -sensing mechanism and H 2 O 2 -dependent structural transition of Corynebacterium glutamicum OxyR by capturing the reduced and H 2 O 2 -bound structures of a serine mutant of the peroxidatic cysteine, and the full-length crystal structure of disulfide-bonded oxidized OxyR. In the H 2 O 2 -bound structure, we pinpoint the key residues for the peroxidatic reduction of H 2 O 2 , and relate this to mutational assays showing that the conserved active-site residues T107 and R278 are critical for effective H 2 O 2 reduction. Furthermore, we propose an allosteric mode of structural change, whereby a localized conformational change arising from H 2 O 2 -induced intramolecular disulfide formation drives a structural shift at the dimerization interface of OxyR, leading to overall changes in quaternary structure and an altered DNA-binding topology and affinity at the catalase promoter region. This study provides molecular insights into the overall OxyR transcription mechanism regulated by H 2 O 2 .
Organizational Affiliation:
Center for Structural Biology, Vlaams Instituut voor Biotechnologie-Vrije Universiteit Brussel, B-1050 Brussels, Belgium.