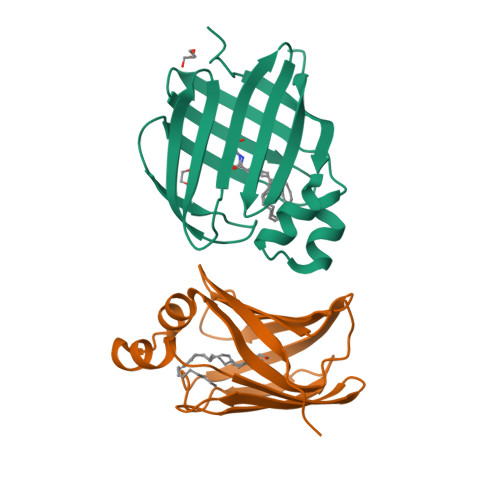
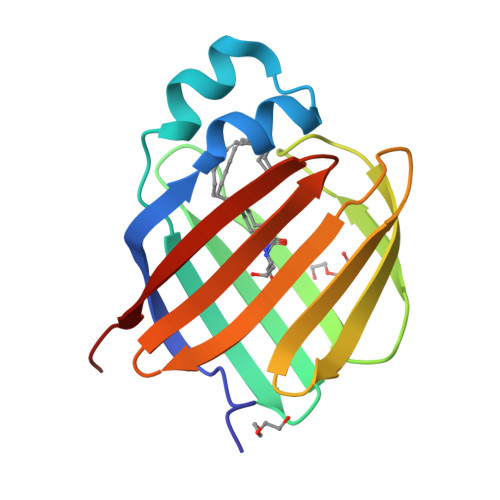
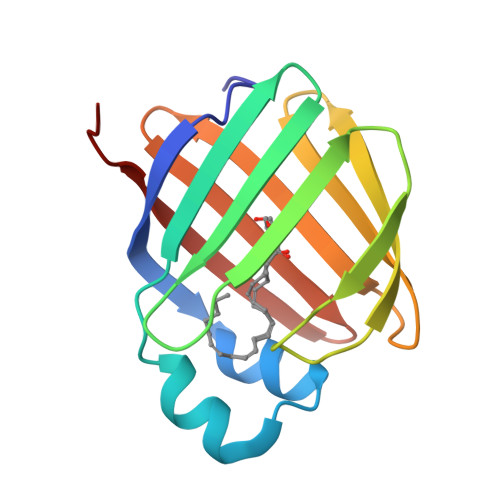
Retinol-binding protein 2 (RBP2) binds monoacylglycerols and modulates gut endocrine signaling and body weight.
Lee, S.A., Yang, K.J.Z., Brun, P.J., Silvaroli, J.A., Yuen, J.J., Shmarakov, I., Jiang, H., Feranil, J.B., Li, X., Lackey, A.I., Krezel, W., Leibel, R.L., Libien, J., Storch, J., Golczak, M., Blaner, W.S.(2020) Sci Adv 6: eaay8937-eaay8937
- PubMed: 32195347
- DOI: https://doi.org/10.1126/sciadv.aay8937
- Primary Citation of Related Structures:
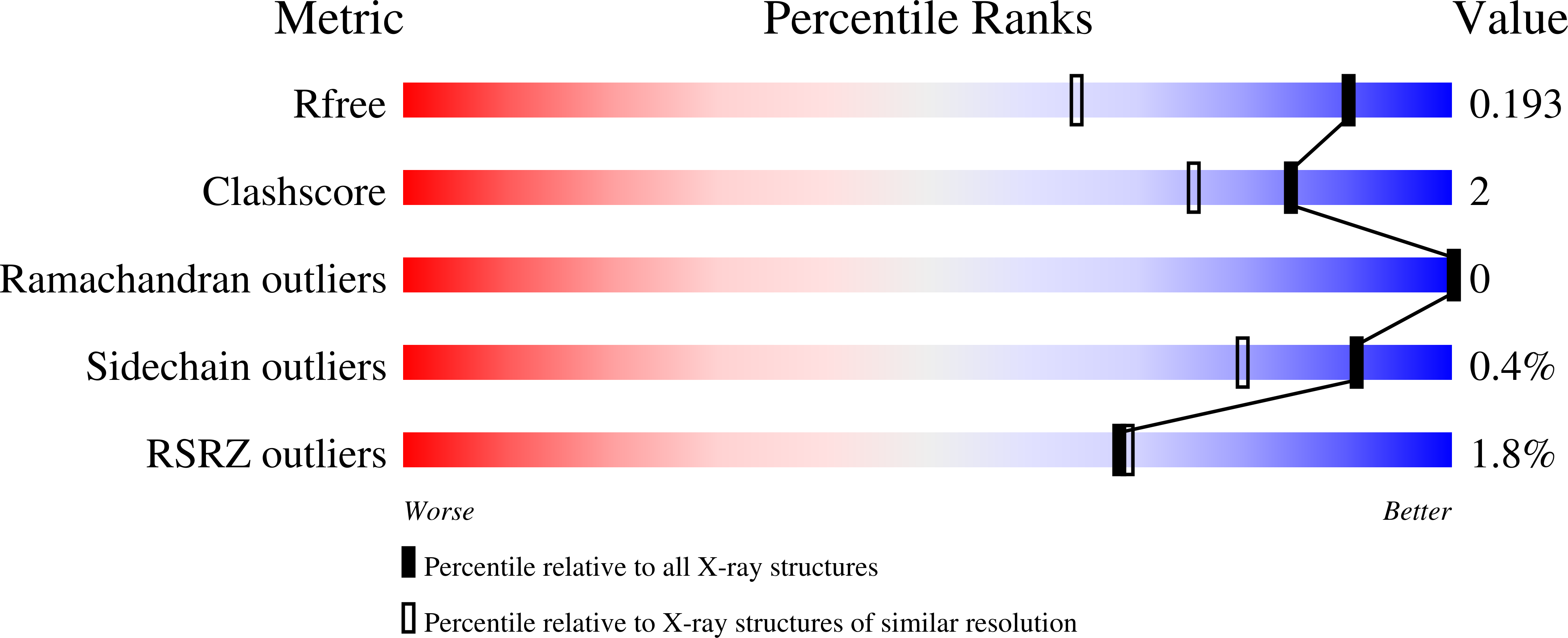
6BTH, 6BTI - PubMed Abstract:
Expressed in the small intestine, retinol-binding protein 2 (RBP2) facilitates dietary retinoid absorption. Rbp2 -deficient ( Rbp2 -/- ) mice fed a chow diet exhibit by 6-7 months-of-age higher body weights, impaired glucose metabolism, and greater hepatic triglyceride levels compared to controls. These phenotypes are also observed when young Rbp2 -/- mice are fed a high fat diet. Retinoids do not account for the phenotypes. Rather, RBP2 is a previously unidentified monoacylglycerol (MAG)-binding protein, interacting with the endocannabinoid 2-arachidonoylglycerol (2-AG) and other MAGs with affinities comparable to retinol. X-ray crystallographic studies show that MAGs bind in the retinol binding pocket. When challenged with an oil gavage, Rbp2 -/- mice show elevated mucosal levels of 2-MAGs. This is accompanied by significantly elevated blood levels of the gut hormone GIP (glucose-dependent insulinotropic polypeptide). Thus, RBP2, in addition to facilitating dietary retinoid absorption, modulates MAG metabolism and likely signaling, playing a heretofore unknown role in systemic energy balance.
Organizational Affiliation:
Department of Medicine, Institute of Human Nutrition, College of Physicians and Surgeons, Columbia University, New York, NY, USA.