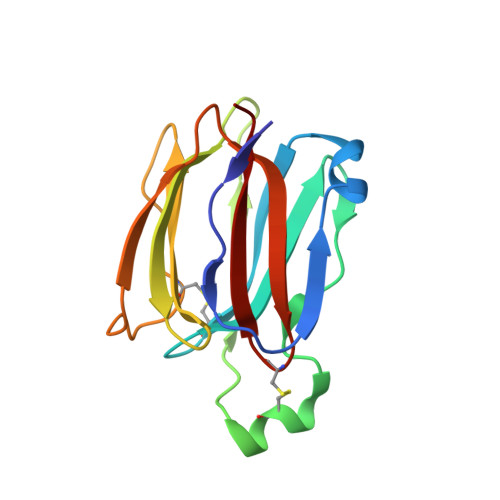
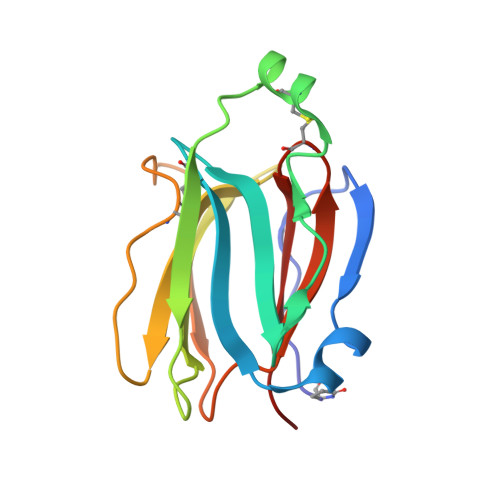
Structures of jacalin-related lectin PPL3 regulating pearl shell biomineralization
Nakae, S., Shionyu, M., Ogawa, T., Shirai, T.(2018) Proteins 86: 644-653
- PubMed: 29524263
- DOI: https://doi.org/10.1002/prot.25491
- Primary Citation of Related Structures:
5YRE, 5YRF, 5YRG, 5YRH, 5YRI, 5YRJ, 5YRK, 5YRL, 5YRM - PubMed Abstract:
The nacreous layer of pearl oysters is one of the major biominerals of commercial and industrial interest. Jacalin-related lectins, including PPL3 isoforms, are known to regulate biomineralization of the Pteria penguin pearl shell, although the molecular mechanisms are largely unknown. The PPL3 crystal structures were determined partly by utilizing microgravity environments for 3 isoforms, namely, PPL3A, PPL3B, and PPL3C. The structures revealed a tail-to-tail dimer structure established by forming a unique inter-subunit disulfide bond at C-termini. The N-terminal residues were found in pyroglutamate form, and this was partly explained by the post-translational modification of PPL3 isoforms implied from the discrepancy between amino acid and gene sequences. The complex structures with trehalose and isomaltose indicated that the novel specificity originated from the unique α-helix of PPL3 isoforms. Docking simulations of PPL3B to various calcite crystal faces suggested the edge of a β-sheet and the carbohydrate-binding site rich in charged residues were the interface to the biomineral, and implied that the isoforms differed in calcite interactions.
- Faculty of Bioscience, Nagahama Institute of Bio-Science and Technology, 1266 Tamura-Cho, Nagahama, Shiga 526-0829, Japan.
Organizational Affiliation: