TOG-tubulin binding specificity promotes microtubule dynamics and mitotic spindle formation.
Byrnes, A.E., Slep, K.C.(2017) J Cell Biol 216: 1641-1657
- PubMed: 28512144
- DOI: https://doi.org/10.1083/jcb.201610090
- Primary Citation of Related Structures:
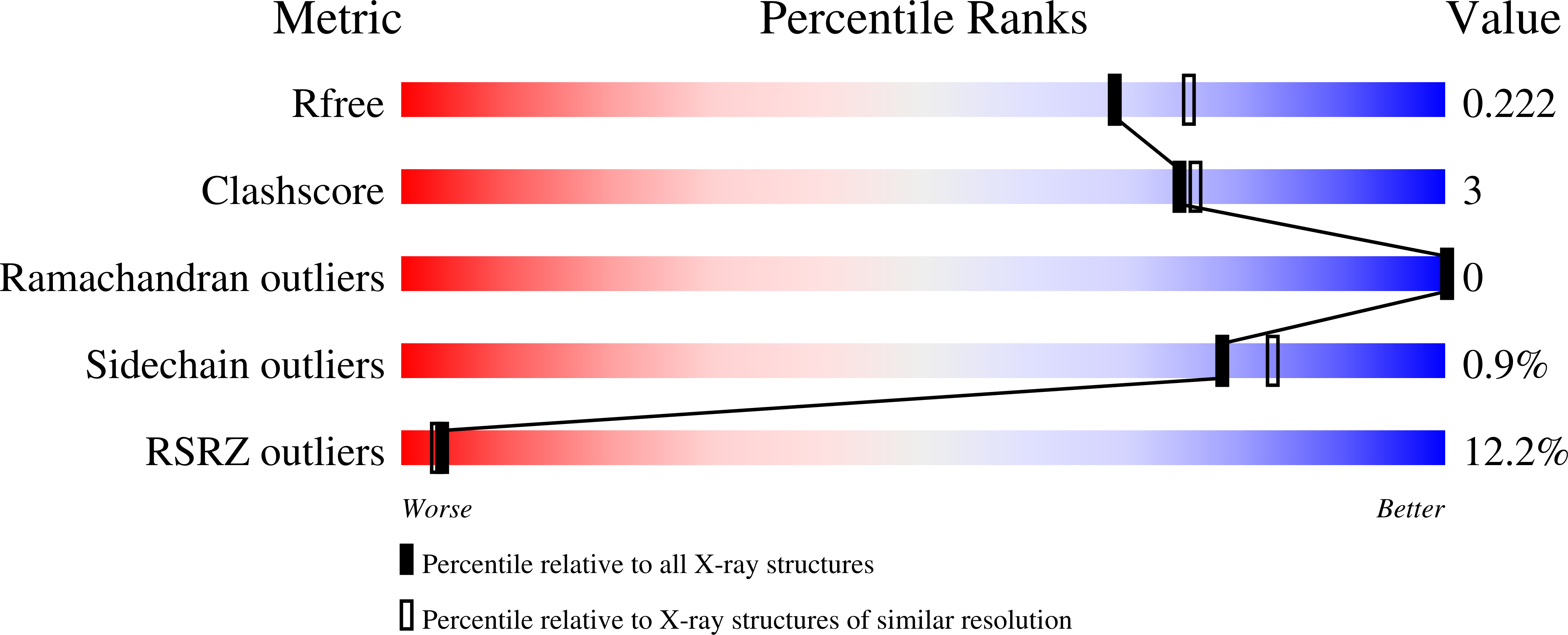
5VJC - PubMed Abstract:
XMAP215, CLASP, and Crescerin use arrayed tubulin-binding tumor overexpressed gene (TOG) domains to modulate microtubule dynamics. We hypothesized that TOGs have distinct architectures and tubulin-binding properties that underlie each family's ability to promote microtubule polymerization or pause. As a model, we investigated the pentameric TOG array of a Drosophila melanogaster XMAP215 member, Msps. We found that Msps TOGs have distinct architectures that bind either free or polymerized tubulin, and that a polarized array drives microtubule polymerization. An engineered TOG1-2-5 array fully supported Msps-dependent microtubule polymerase activity. Requisite for this activity was a TOG5-specific N-terminal HEAT repeat that engaged microtubule lattice-incorporated tubulin. TOG5-microtubule binding maintained mitotic spindle formation as deleting or mutating TOG5 compromised spindle architecture and increased the mitotic index. Mad2 knockdown released the spindle assembly checkpoint triggered when TOG5-microtubule binding was compromised, indicating that TOG5 is essential for spindle function. Our results reveal a TOG5-specific role in mitotic fidelity and support our hypothesis that architecturally distinct TOGs arranged in a sequence-specific order underlie TOG array microtubule regulator activity.
Organizational Affiliation:
Department of Biochemistry and Biophysics, University of North Carolina, Chapel Hill, NC 27599.