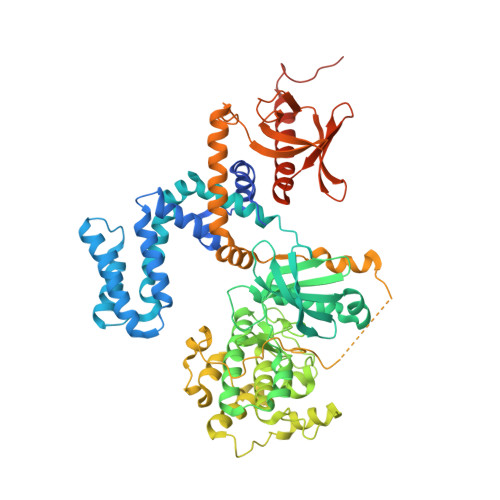
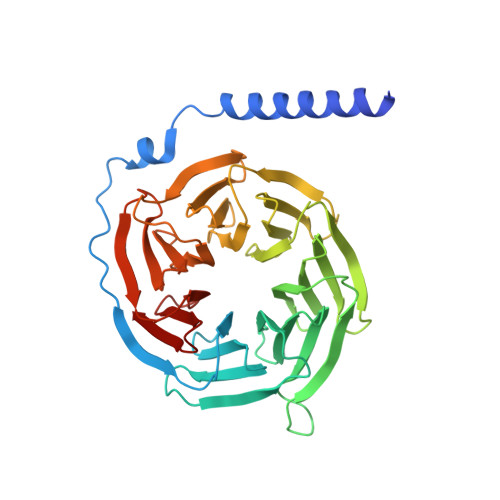
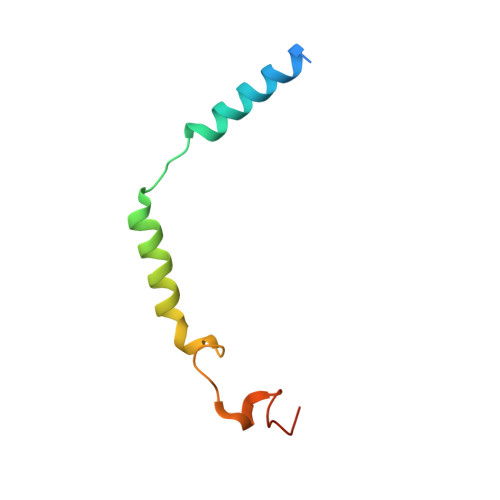
Structure-Based Design of Highly Selective and Potent G Protein-Coupled Receptor Kinase 2 Inhibitors Based on Paroxetine.
Waldschmidt, H.V., Homan, K.T., Cato, M.C., Cruz-Rodriguez, O., Cannavo, A., Wilson, M.W., Song, J., Cheung, J.Y., Koch, W.J., Tesmer, J.J., Larsen, S.D.(2017) J Med Chem 60: 3052-3069
- PubMed: 28323425
- DOI: https://doi.org/10.1021/acs.jmedchem.7b00112
- Primary Citation of Related Structures:
5UKK, 5UKL, 5UKM - PubMed Abstract:
In heart failure, the β-adrenergic receptors (βARs) become desensitized and uncoupled from heterotrimeric G proteins. This process is initiated by G protein-coupled receptor kinases (GRKs), some of which are upregulated in the failing heart, making them desirable therapeutic targets. The selective serotonin reuptake inhibitor, paroxetine, was previously identified as a GRK2 inhibitor. Utilizing a structure-based drug design approach, we modified paroxetine to generate a small compound library. Included in this series is a highly potent and selective GRK2 inhibitor, 14as, with an IC 50 of 30 nM against GRK2 and greater than 230-fold selectivity over other GRKs and kinases. Furthermore, 14as showed a 100-fold improvement in cardiomyocyte contractility assays over paroxetine and a plasma concentration higher than its IC 50 for over 7 h. Three of these inhibitors, including 14as, were additionally crystallized in complex with GRK2 to give insights into the structural determinants of potency and selectivity of these inhibitors.
Organizational Affiliation:
Department of Medicinal Chemistry, College of Pharmacy, ‡Departments of Pharmacology and Biological Chemistry, Life Sciences Institute, §Ph.D. Program in Chemical Biology, ⊥Vahlteich Medicinal Chemistry Core, University of Michigan , Ann Arbor, Michigan 48109, United States.