ZATT (ZNF451)-mediated resolution of topoisomerase 2 DNA-protein cross-links.
Schellenberg, M.J., Lieberman, J.A., Herrero-Ruiz, A., Butler, L.R., Williams, J.G., Munoz-Cabello, A.M., Mueller, G.A., London, R.E., Cortes-Ledesma, F., Williams, R.S.(2017) Science 357: 1412-1416
- PubMed: 28912134
- DOI: https://doi.org/10.1126/science.aam6468
- Primary Citation of Related Structures:
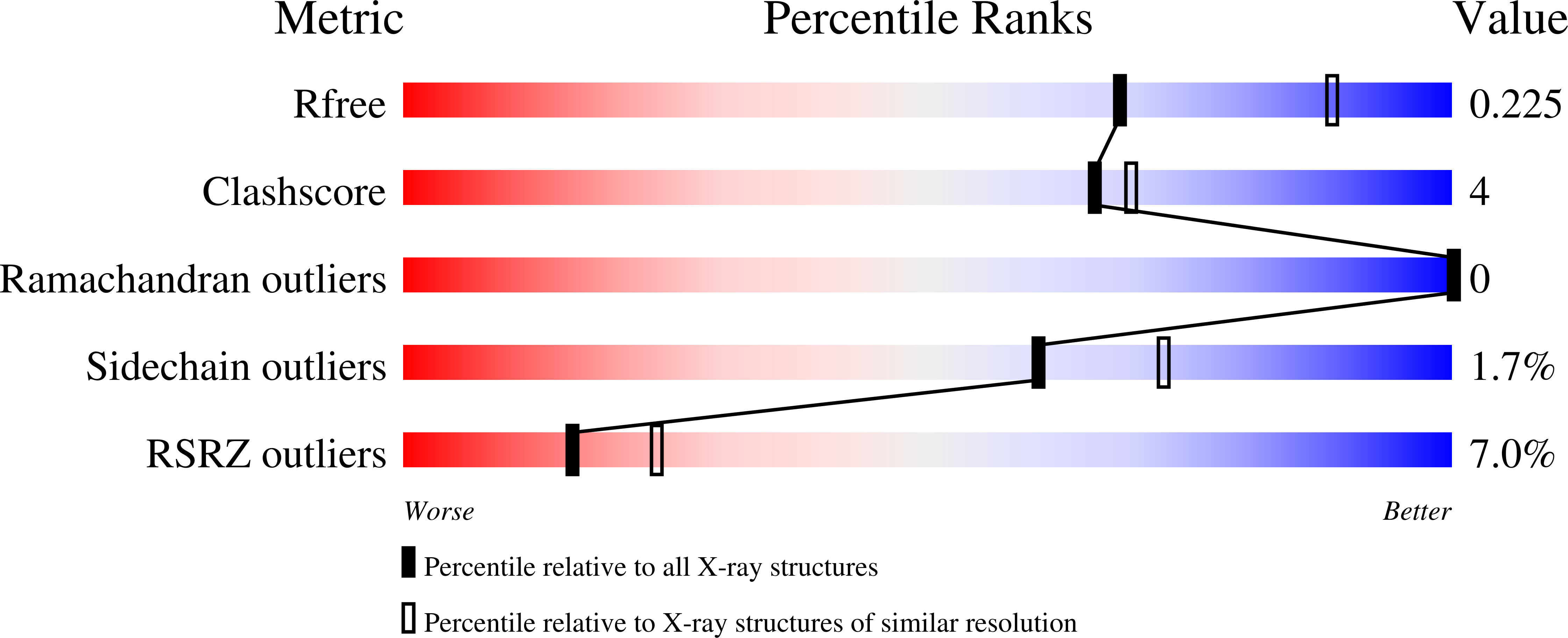
5TVP, 5TVQ - PubMed Abstract:
Topoisomerase 2 (TOP2) DNA transactions proceed via formation of the TOP2 cleavage complex (TOP2cc), a covalent enzyme-DNA reaction intermediate that is vulnerable to trapping by potent anticancer TOP2 drugs. How genotoxic TOP2 DNA-protein cross-links are resolved is unclear. We found that the SUMO (small ubiquitin-related modifier) ligase ZATT (ZNF451) is a multifunctional DNA repair factor that controls cellular responses to TOP2 damage. ZATT binding to TOP2cc facilitates a proteasome-independent tyrosyl-DNA phosphodiesterase 2 (TDP2) hydrolase activity on stalled TOP2cc. The ZATT SUMO ligase activity further promotes TDP2 interactions with SUMOylated TOP2, regulating efficient TDP2 recruitment through a "split-SIM" SUMO2 engagement platform. These findings uncover a ZATT-TDP2-catalyzed and SUMO2-modulated pathway for direct resolution of TOP2cc.
Organizational Affiliation:
Genome Integrity and Structural Biology Laboratory, National Institute of Environmental Health Sciences (NIEHS), Research Triangle Park, NC 27709, USA.