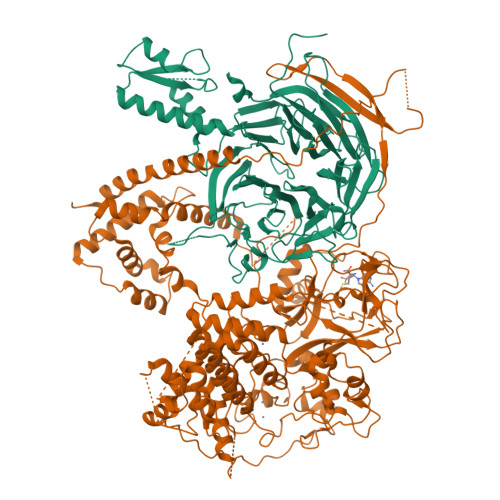
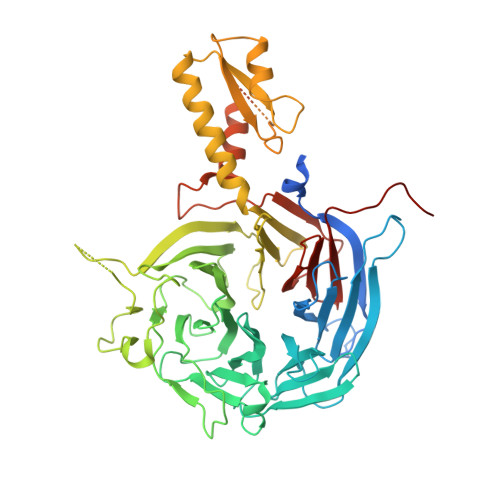
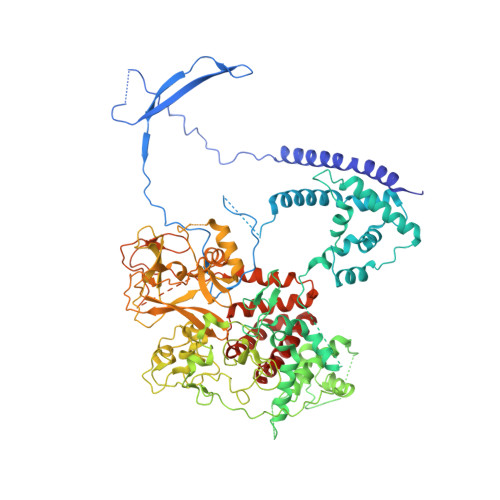
Polycomb repressive complex 2 in an autoinhibited state.
Bratkowski, M., Yang, X., Liu, X.(2017) J Biological Chem 292: 13323-13332
- PubMed: 28607149
- DOI: https://doi.org/10.1074/jbc.M117.787572
- Primary Citation of Related Structures:
5BJS, 5TQR, 5VK3 - PubMed Abstract:
Polycomb-group proteins control many fundamental biological processes, such as anatomical development in mammals and vernalization in plants. Polycomb repressive complex 2 (PRC2) is responsible for methylation of histone H3 lysine 27 (H3K27), and trimethylated H3K27 (H3K27me3) is implicated in epigenetic gene silencing. Recent genomic, biochemical, and structural data indicate that PRC2 is broadly conserved from yeast to human in many aspects. Here, we determined the crystal structure of an apo-PRC2 from the fungus Chaetomium thermophilum captured in a bona fide autoinhibited state, which represents a novel conformation of PRC2 associated with enzyme regulation in light of the basal and stimulated states that we reported previously. We found that binding by the cofactor S -adenosylmethionine mitigates this autoinhibited structural state. Using steady-state enzyme kinetics, we also demonstrated that disrupting the autoinhibition results in a vastly activated enzyme complex. Autoinhibition provides a novel structural platform that may enable control of PRC2 activity in response to diverse transcriptional states and chromatin contexts and set a ground state to allow PRC2 activation by other cellular mechanisms as well.
Organizational Affiliation:
From the Cecil H. and Ida Green Center for Reproductive Biology Sciences and Division of Basic Research, Department of Obstetrics and Gynecology, Department of Biophysics, UT Southwestern Medical Center, Dallas, Texas 75390.