Characterization of a drug-targetable allosteric site regulating vascular endothelial growth factor signaling.
Thieltges, K.M., Avramovic, D., Piscitelli, C.L., Markovic-Mueller, S., Binz, H.K., Ballmer-Hofer, K.(2018) Angiogenesis 21: 533-543
- PubMed: 29502220
- DOI: https://doi.org/10.1007/s10456-018-9606-9
- Primary Citation of Related Structures:
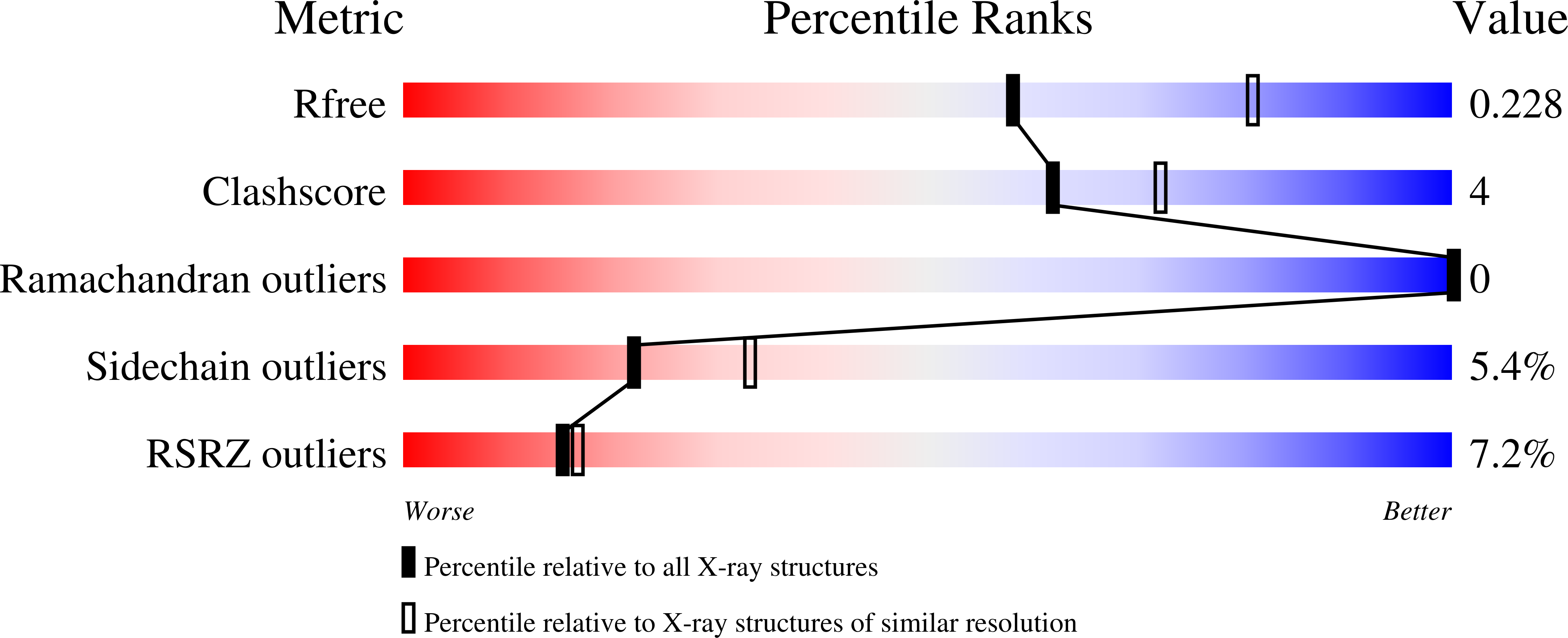
5OYJ - PubMed Abstract:
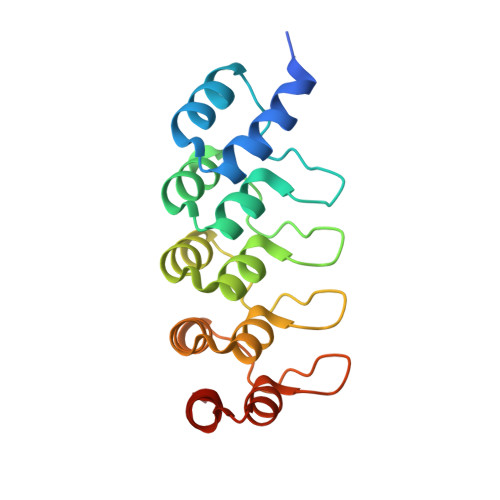
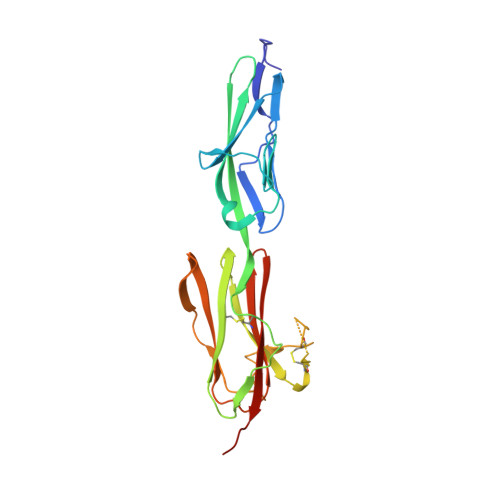
Vascular endothelial growth factors (VEGFs) regulate blood and lymph vessel development upon activation of three receptor tyrosine kinases (VEGFRs). The extracellular domain of VEGFRs consists of seven Ig-homology domains, of which D2-3 form the ligand-binding site, while the membrane proximal domains D4-7 are involved in homotypic interactions in ligand-bound receptor dimers. Based on low-resolution structures, we identified allosteric sites in D4-5 and D7 of vascular endothelial growth factor receptor 2 (VEGFR-2) accomplishing regulatory functions. Allosteric inhibition of VEGFR-2 signaling represents an attractive option for the treatment of neovascular diseases. We showed earlier that DARPin ® binders to domains D4 or D7 are potent VEGFR-2 inhibitors. Here we investigated in detail the allosteric inhibition mechanism of the domain D4 binding inhibitor D4b. The 2.38 Å crystal structure of D4b in complex with VEGFR-2 D4-5, the first high-resolution structure of this VEGFR-2 segment, indicates steric hindrance by D4b as the mechanism of inhibition of receptor activation. At the cellular level, D4b triggered quantitative internalization of VEGFR-2 in the absence of ligand and thus clearance of VEGFR-2 from the surface of endothelial cells. The allosteric VEGFR-2 inhibition was sufficiently strong to efficiently inhibit the growth of human endothelial cells at suboptimal dose in a mouse xenograft model in vivo, underlining the therapeutic potential of the approach.
Organizational Affiliation:
Laboratory of Biomolecular Research, Paul Scherrer Institut, 5232, Villigen, Switzerland.