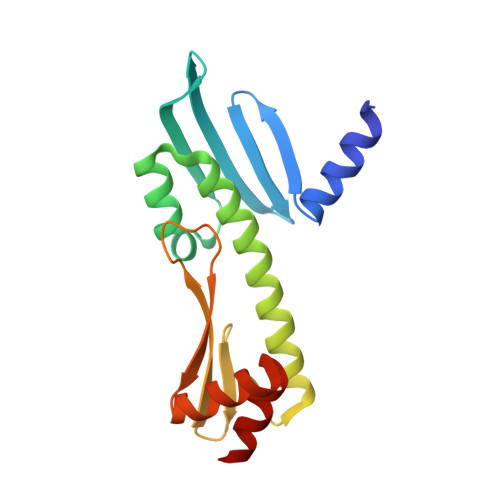
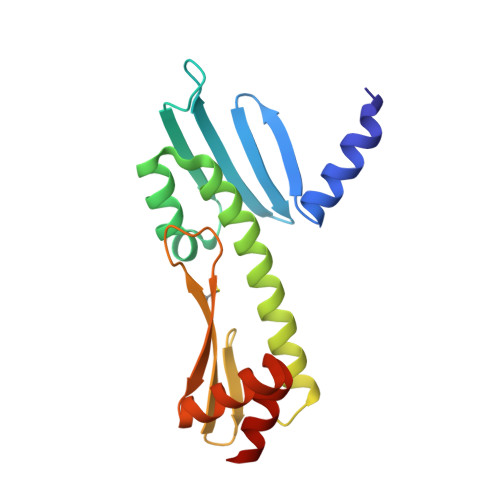
Molecular basis for inner kinetochore configuration through RWD domain-peptide interactions.
Schmitzberger, F., Richter, M.M., Gordiyenko, Y., Robinson, C.V., Dadlez, M., Westermann, S.(2017) EMBO J 36: 3458-3482
- PubMed: 29046335
- DOI: https://doi.org/10.15252/embj.201796636
- Primary Citation of Related Structures:
5MU3 - PubMed Abstract:
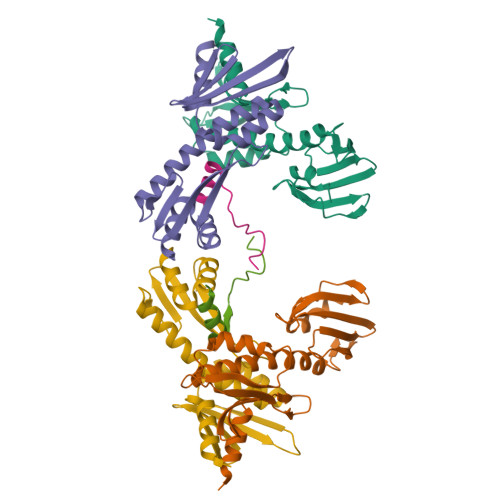
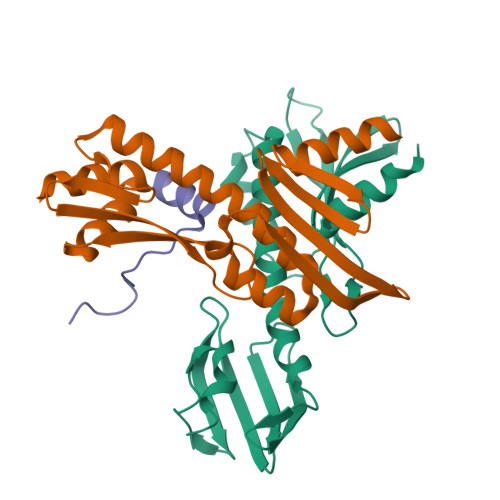
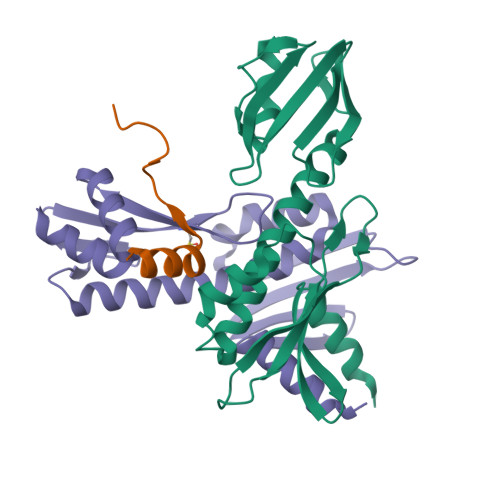
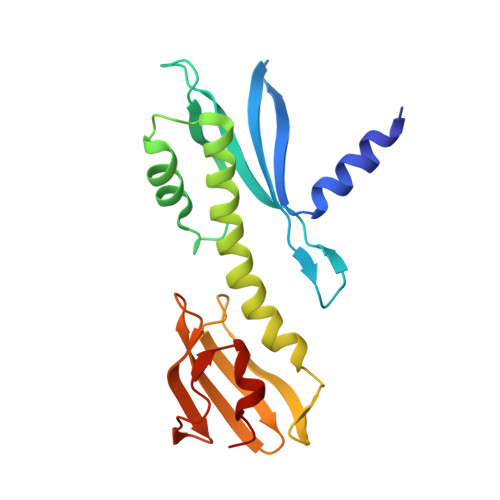
Kinetochores are dynamic cellular structures that connect chromosomes to microtubules. They form from multi-protein assemblies that are evolutionarily conserved between yeasts and humans. One of these assemblies-COMA-consists of subunits Ame1 CENP-U , Ctf19 CENP-P , Mcm21 CENP-O and Okp1 CENP-Q A description of COMA molecular organization has so far been missing. We defined the subunit topology of COMA, bound with inner kinetochore proteins Nkp1 and Nkp2, from the yeast Kluyveromyces lactis , with nanoflow electrospray ionization mass spectrometry, and mapped intermolecular contacts with hydrogen-deuterium exchange coupled to mass spectrometry. Our data suggest that the essential Okp1 subunit is a multi-segmented nexus with distinct binding sites for Ame1, Nkp1-Nkp2 and Ctf19-Mcm21. Our crystal structure of the Ctf19-Mcm21 RWD domains bound with Okp1 shows the molecular contacts of this important inner kinetochore joint. The Ctf19-Mcm21 binding motif in Okp1 configures a branch of mitotic inner kinetochores, by tethering Ctf19-Mcm21 and Chl4 CENP-N -Iml3 CENP-L Absence of this motif results in dependence on the mitotic checkpoint for viability.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA, USA florian.schmitzberger@uni-due.de.