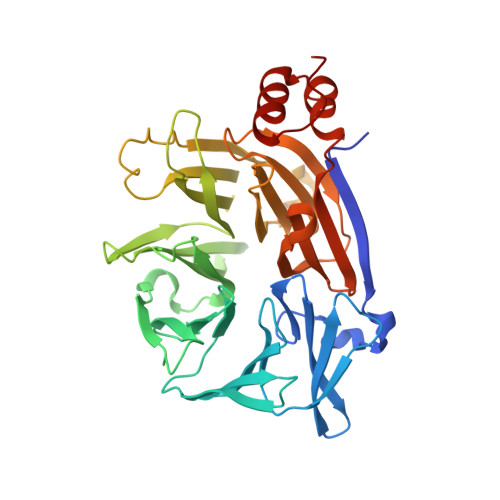
Cellular and viral peptides bind multiple sites on the N-terminal domain of clathrin.
Muenzner, J., Traub, L.M., Kelly, B.T., Graham, S.C.(2017) Traffic 18: 44-57
- PubMed: 27813245
- DOI: https://doi.org/10.1111/tra.12457
- Primary Citation of Related Structures:
5M5R, 5M5S, 5M5T, 5M5U, 5M5V, 5M61 - PubMed Abstract:
Short peptide motifs in unstructured regions of clathrin-adaptor proteins recruit clathrin to membranes to facilitate post-Golgi membrane transport. Three consensus clathrin-binding peptide sequences have been identified and structural studies show that each binds distinct sites on the clathrin heavy chain N-terminal domain (NTD). A fourth binding site for adaptors on NTD has been functionally identified but not structurally characterised. We have solved high resolution structures of NTD bound to peptide motifs from the cellular clathrin adaptors β2 adaptin and amphiphysin plus a putative viral clathrin adaptor, hepatitis D virus large antigen (HDAg-L). Surprisingly, with each peptide we observe simultaneous peptide binding at multiple sites on NTD and viral peptides binding to the same sites as cellular peptides. Peptides containing clathrin-box motifs (CBMs) with the consensus sequence LΦxΦ[DE] bind at the 'arrestin box' on NTD, between β-propeller blades 4 and 5, which had previously been thought to bind a distinct consensus sequence. Further, we structurally define the fourth peptide binding site on NTD, which we term the Royle box. In vitro binding assays show that clathrin is more readily captured by cellular CBMs than by HDAg-L, and site-directed mutagenesis confirms that multiple binding sites on NTD contribute to efficient capture by CBM peptides.
Organizational Affiliation:
Department of Pathology, University of Cambridge, Cambridge, UK.