Inflammation-Induced Adhesin-Receptor Interaction Provides a Fitness Advantage to Uropathogenic E. coli during Chronic Infection.
Conover, M.S., Ruer, S., Taganna, J., Kalas, V., De Greve, H., Pinkner, J.S., Dodson, K.W., Remaut, H., Hultgren, S.J.(2016) Cell Host Microbe 20: 482-492
- PubMed: 27667696
- DOI: https://doi.org/10.1016/j.chom.2016.08.013
- Primary Citation of Related Structures:
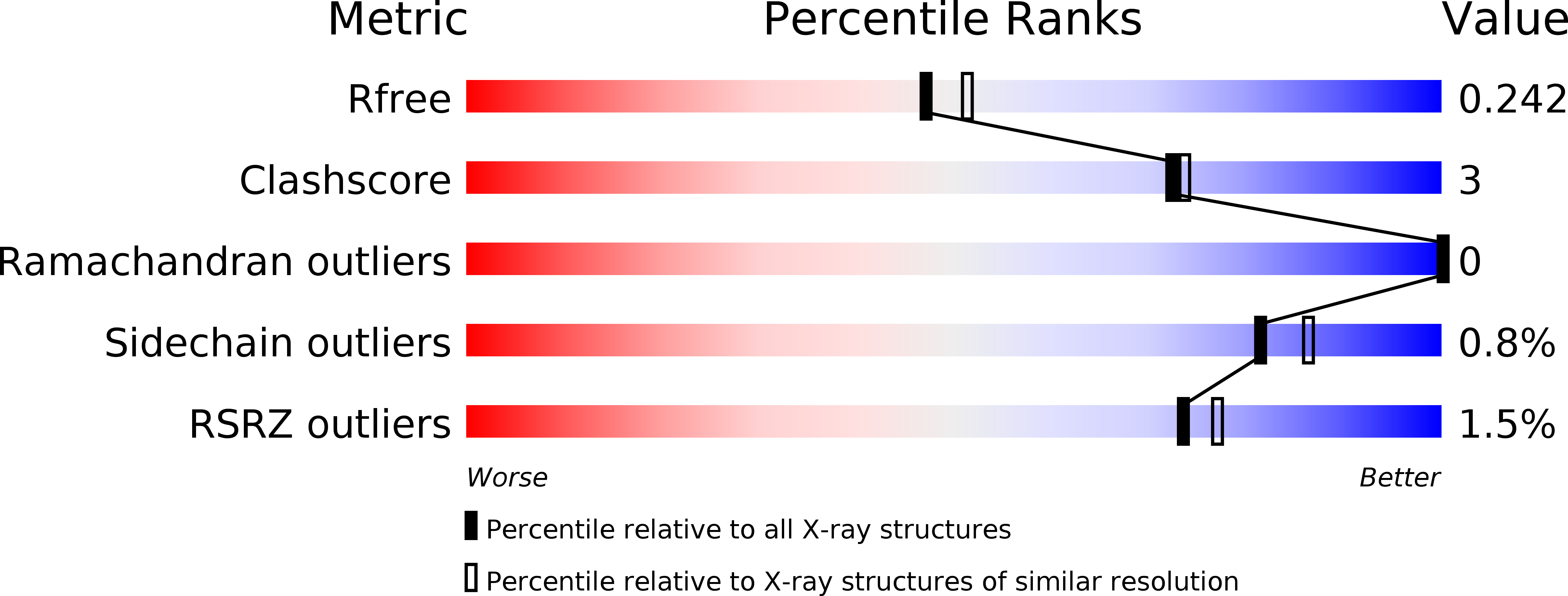
5LNE, 5LNG - PubMed Abstract:
Uropathogenic E. coli (UPEC) is the dominant cause of urinary tract infections, clinically described as cystitis. UPEC express CUP pili, which are extracellular fibers tipped with adhesins that bind mucosal surfaces of the urinary tract. Here we identify the role of the F9/Yde/Fml pilus for UPEC persistence in the inflamed urothelium. The Fml adhesin FmlH binds galactose β1-3 N-acetylgalactosamine found in core-1 and -2 O-glycans. Deletion of fmlH had no effect on UPEC virulence in an acute mouse model of cystitis. However, FmlH provided a fitness advantage during chronic cystitis, which is manifested as persistent bacteriuria, high bladder bacterial burdens, and chronic inflammation. In situ binding confirmed that FmlH bound avidly to the inflamed, but not the naive bladder. In accordance with its pathogenic profile, vaccination with FmlH significantly protected mice from chronic cystitis. Thus, UPEC employ separate CUP pili to adapt to the rapidly changing niche during bladder infection.
Organizational Affiliation:
Department of Molecular Microbiology and Center for Women's Infectious Disease Research, Washington University School of Medicine, St. Louis, MO 63110, USA.