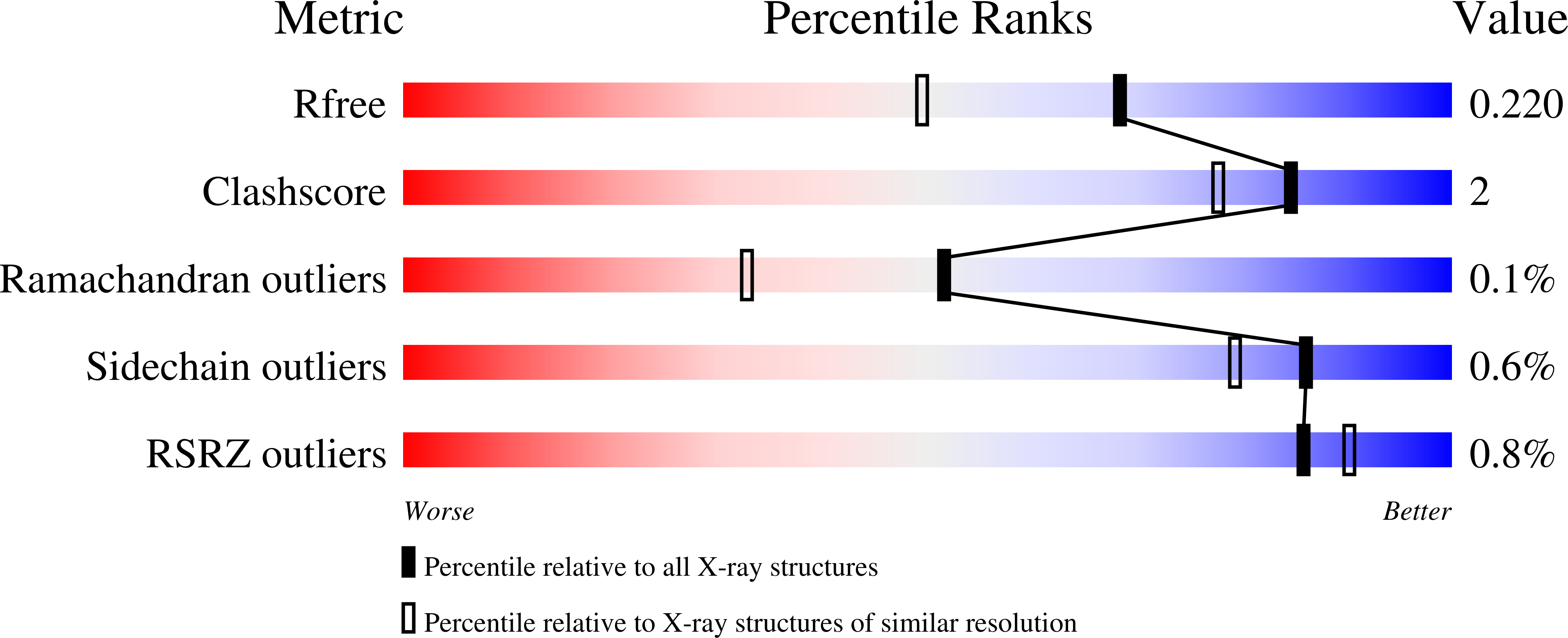
Crystal structure of a peptidyl-dipeptidase K-26-DCP from Actinomycete in complex with its natural inhibitor.
Masuyer, G., Cozier, G.E., Kramer, G.J., Bachmann, B.O., Acharya, K.R.(2016) FEBS J 283: 4357-4369
- PubMed: 27754586
- DOI: https://doi.org/10.1111/febs.13928
- Primary Citation of Related Structures:
5L43, 5L44 - PubMed Abstract:
Several soil-derived Actinobacteria produce secondary metabolites that are proven specific and potent inhibitors of the human angiotensin-I-converting enzyme (ACE), a key target for the modulation of hypertension through its role in the renin-angiotensin-aldosterone system. K-26-DCP is a zinc dipeptidyl carboxypeptidase (DCP) produced by Astrosporangium hypotensionis, and an ancestral homologue of ACE. Here we report the high-resolution crystal structures of K-26-DCP and of its complex with the natural microbial tripeptide product K-26. The experimental results provide the structural basis for better understanding the specificity of K-26 for human ACE over bacterial DCPs. Structural data are available in the PDB under the accession numbers 5L43 and 5L44.
Organizational Affiliation:
Department of Biology and Biochemistry, University of Bath, UK.