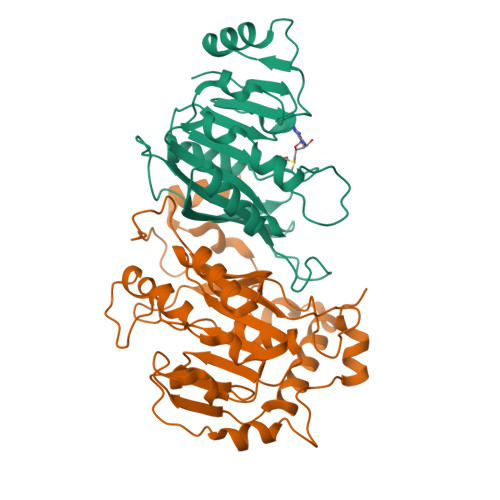
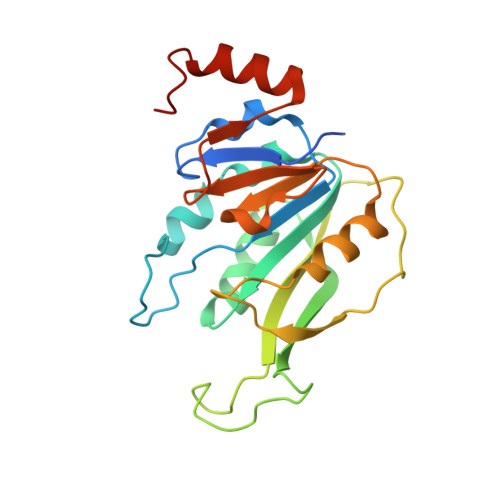
Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases.
Wang, P., Doxtader, K.A., Nam, Y.(2016) Mol Cell 63: 306-317
- PubMed: 27373337
- DOI: https://doi.org/10.1016/j.molcel.2016.05.041
- Primary Citation of Related Structures:
5K7M, 5K7U, 5K7W - PubMed Abstract:
N(6)-methyladenosine (m(6)A) is a prevalent, reversible chemical modification of functional RNAs and is important for central events in biology. The core m(6)A writers are Mettl3 and Mettl14, which both contain methyltransferase domains. How Mettl3 and Mettl14 cooperate to catalyze methylation of adenosines has remained elusive. We present crystal structures of the complex of Mettl3/Mettl14 methyltransferase domains in apo form as well as with bound S-adenosylmethionine (SAM) or S-adenosylhomocysteine (SAH) in the catalytic site. We determine that the heterodimeric complex of methyltransferase domains, combined with CCCH motifs, constitutes the minimally required regions for creating m(6)A modifications in vitro. We also show that Mettl3 is the catalytically active subunit, while Mettl14 plays a structural role critical for substrate recognition. Our model provides a molecular explanation for why certain mutations of Mettl3 and Mettl14 lead to impaired function of the methyltransferase complex.
Organizational Affiliation:
Cecil H. and Ida Green Center for Reproductive Biology Sciences, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA; Division of Basic Reproductive Biology Research, Department of Obstetrics and Gynecology, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA; Department of Biophysics, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA.