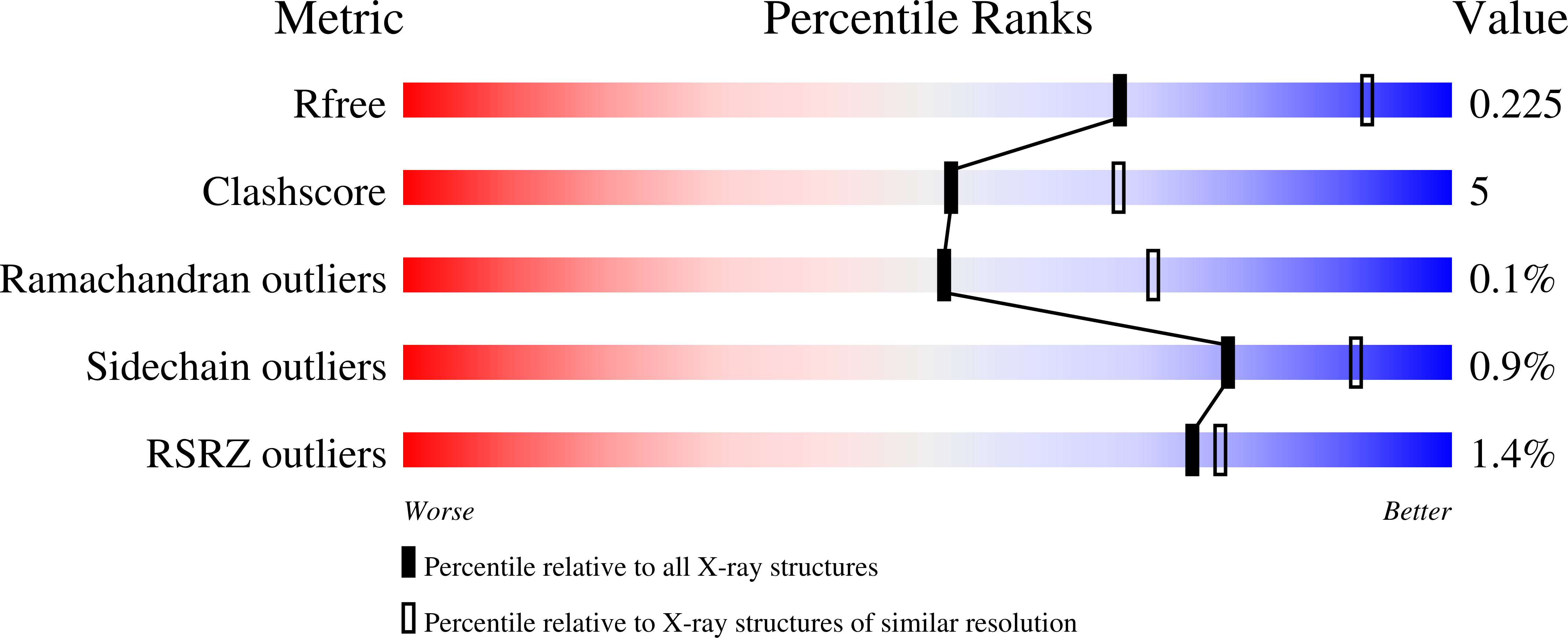
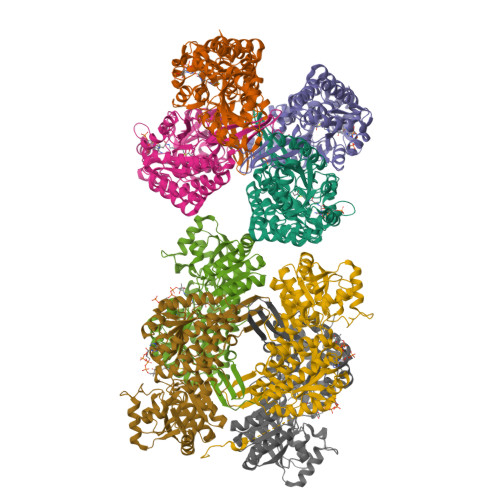
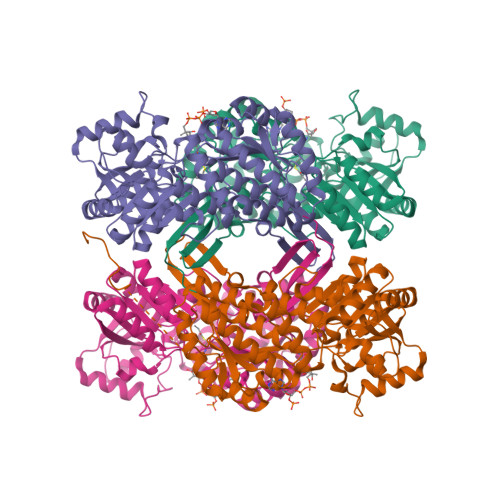
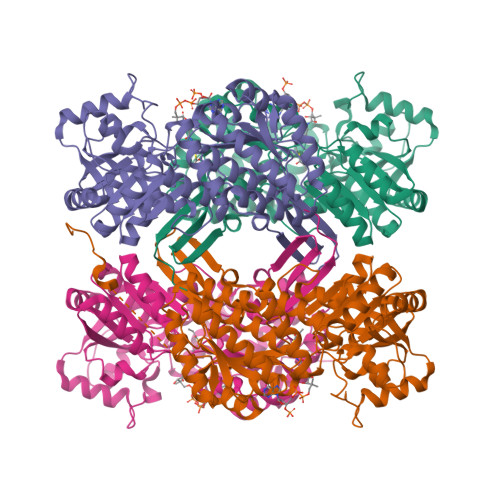
In Vitro Characterization and Concerted Function of Three Core Enzymes of a Glycyl Radical Enzyme - Associated Bacterial Microcompartment.
Zarzycki, J., Sutter, M., Cortina, N.S., Erb, T.J., Kerfeld, C.A.(2017) Sci Rep 7: 42757-42757
- PubMed: 28202954
- DOI: https://doi.org/10.1038/srep42757
- Primary Citation of Related Structures:
5JFL, 5JFM, 5JFN - PubMed Abstract:
Many bacteria encode proteinaceous bacterial microcompartments (BMCs) that encapsulate sequential enzymatic reactions of diverse metabolic pathways. Well-characterized BMCs include carboxysomes for CO 2 -fixation, and propanediol- and ethanolamine-utilizing microcompartments that contain B 12 -dependent enzymes. Genes required to form BMCs are typically organized in gene clusters, which promoted their distribution across phyla by horizontal gene transfer. Recently, BMCs associated with glycyl radical enzymes (GREs) were discovered; these are widespread and comprise at least three functionally distinct types. Previously, we predicted one type of these GRE-associated microcompartments (GRMs) represents a B 12 -independent propanediol-utilizing BMC. Here we functionally and structurally characterize enzymes of the GRM of Rhodopseudomonas palustris BisB18 and demonstrate their concerted function in vitro. The GRM signature enzyme, the GRE, is a dedicated 1,2-propanediol dehydratase with a new type of intramolecular encapsulation peptide. It forms a complex with its activating enzyme and, in conjunction with an aldehyde dehydrogenase, converts 1,2-propanediol to propionyl-CoA. Notably, homologous GRMs are also encoded in pathogenic Escherichia coli strains. Our high-resolution crystal structures of the aldehyde dehydrogenase lead to a revised reaction mechanism. The successful in vitro reconstitution of a part of the GRM metabolism provides insights into the metabolic function and steps in the assembly of this BMC.
Organizational Affiliation:
Max-Planck-Institute for Terrestrial Microbiology, Karl-von-Frisch-Str. 10, D-35043, Marburg, Germany.