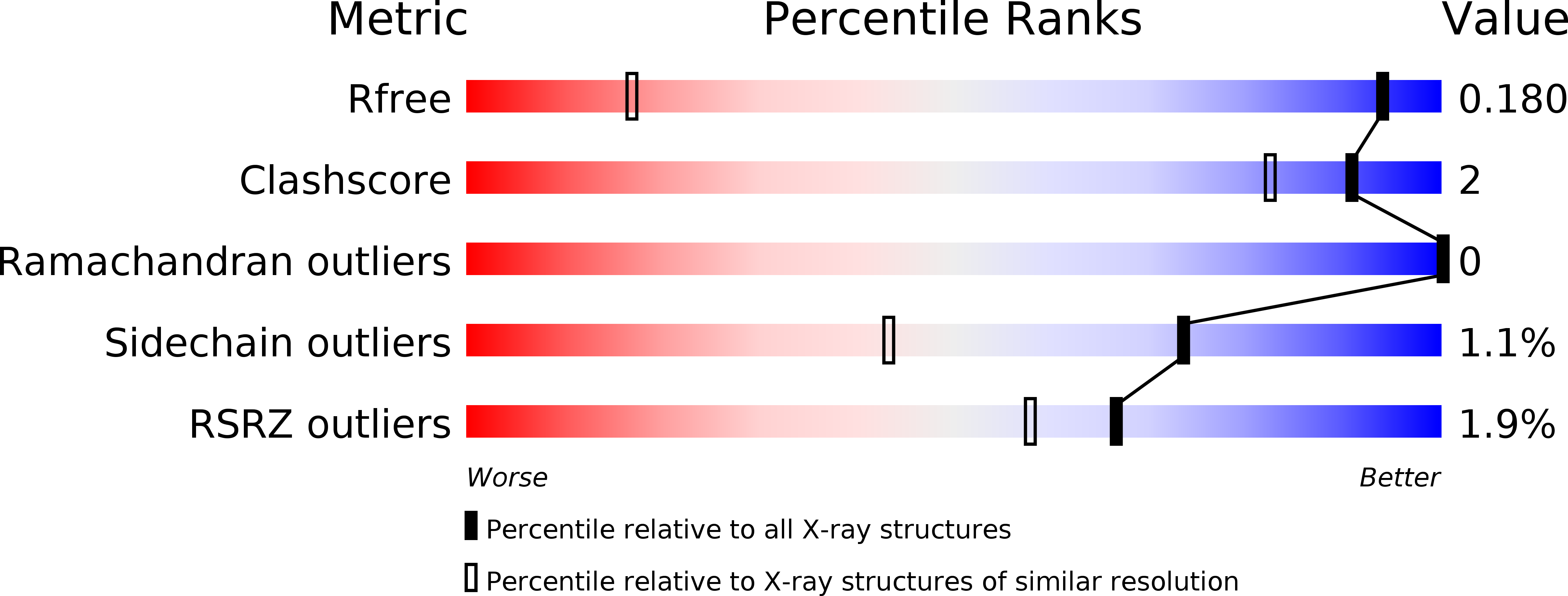
Sap1 is a replication-initiation factor essential for the assembly of pre-replicative complex in the fission yeast Schizosaccharomyces pombe.
Guan, L., He, P., Yang, F., Zhang, Y., Hu, Y., Ding, J., Hua, Y., Zhang, Y., Ye, Q., Hu, J., Wang, T., Jin, C., Kong, D.(2017) J Biol Chem 292: 6056-6075
- PubMed: 28223353
- DOI: https://doi.org/10.1074/jbc.M116.767806
- Primary Citation of Related Structures:
5B7J, 5JDK - PubMed Abstract:
A central step in the initiation of chromosomal DNA replication in eukaryotes is the assembly of pre-replicative complex (pre-RC) at late M and early G 1 phase of the cell cycles. Since 1973, four proteins or protein complexes, including cell division control protein 6 (Cdc6)/Cdc18, minichromosome maintenance protein complex, origin recognition complex (ORC), and Cdt1, are known components of the pre-RC. Previously, we reported that a non-ORC protein binds to the essential element Δ9 of the Schizosaccharomyces pombe DNA-replication origin ARS3001. In this study, we identified that the non-ORC protein is Sap1. Like ORC, Sap1 binds to DNA origins during cell growth cycles. But unlike ORC, which binds to asymmetric AT-rich sequences through its nine AT-hook motifs, Sap1 preferentially binds to a DNA sequence of 5'-(A/T) n (C/G)(A/T) 9-10 (G/C)(A/T) n -3' ( n ≥ 1). We also found that Sap1 and ORC physically interact. We further demonstrated that Sap1 is required for the assembly of the pre-RC because of its essential role in recruiting Cdc18 to DNA origins. Thus, we conclude that Sap1 is a replication-initiation factor that directly participates in the assembly of the pre-RC. DNA-replication origins in fission yeast are defined by possessing two essential elements with one bound by ORC and the other by Sap1.
Organizational Affiliation:
From the Peking-Tsinghua Center for Life Sciences, National Laboratory of Protein and Plant Gene Research, College of Life Sciences.