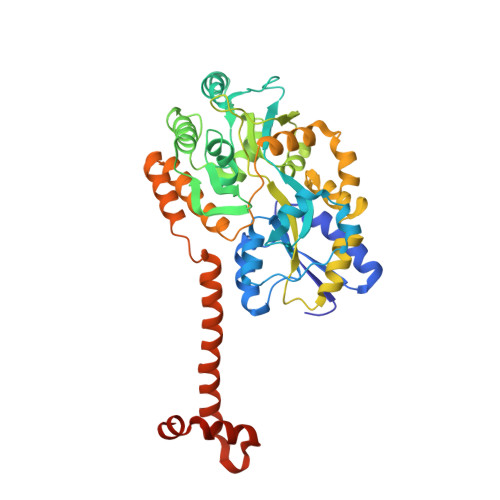
Crystal structure of the N-terminal domain of human SIRT7 reveals a three-helical domain architecture
Priyanka, A., Solanki, V., Parkesh, R., Thakur, K.G.(2016) Proteins 84: 1558-1563
- PubMed: 27287224
- DOI: https://doi.org/10.1002/prot.25085
- Primary Citation of Related Structures:
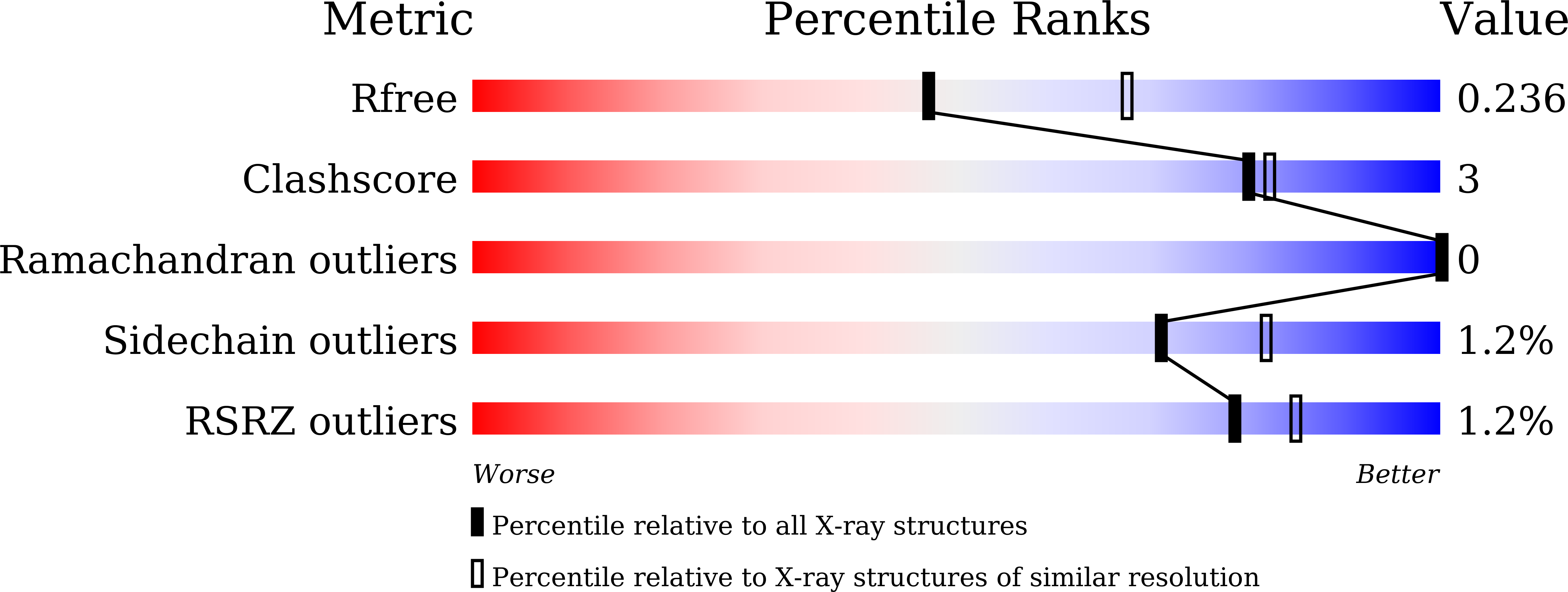
5IQZ - PubMed Abstract:
Human SIRT7 is an NAD(+) dependent deacetylase, which belongs to sirtuin family of proteins. SIRT7, like other sirtuins has conserved catalytic domain and is flanked by N- and C-terminal domains reported to play vital functional roles. Here, we report the crystal structure of the N-terminal domain of human SIRT7 (SIRT7(NTD) ) at 2.3 Å resolution as MBP-SIRT7(NTD) fusion protein. SIRT7(NTD) adopts three-helical domain architecture and comparative structural analyses suggest similarities to some DNA binding motifs and transcription regulators. We also report here the importance of N- and C-terminal domains in soluble expression of SIRT7. Proteins 2016; 84:1558-1563. © 2016 Wiley Periodicals, Inc.
Organizational Affiliation:
Structural Biology Laboratory, G. N. Ramachandran Protein Centre, CSIR-Institute of Microbial Technology, Chandigarh, 160036, India.