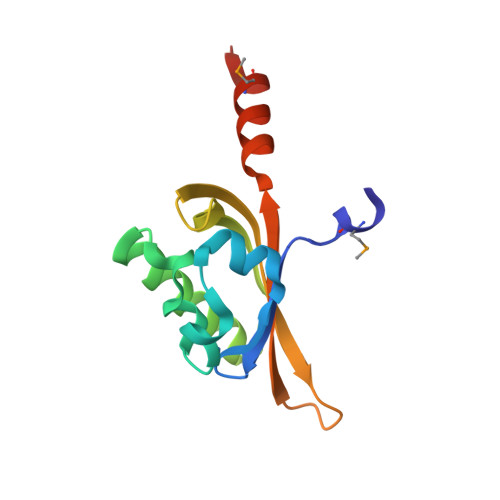
Molecular Insights into Toluene Sensing in the TodS/TodT Signal Transduction System.
Koh, S., Hwang, J., Guchhait, K., Lee, E.G., Kim, S.Y., Kim, S., Lee, S., Chung, J.M., Jung, H.S., Lee, S.J., Ryu, C.M., Lee, S.G., Oh, T.K., Kwon, O., Kim, M.H.(2016) J Biological Chem 291: 8575-8590
- PubMed: 26903514
- DOI: https://doi.org/10.1074/jbc.M116.718841
- Primary Citation of Related Structures:
5HWT, 5HWV, 5HWW - PubMed Abstract:
TodS is a sensor kinase that responds to various monoaromatic compounds, which either cause an agonistic or antagonistic effect on phosphorylation of its cognate response regulator TodT, and controls tod operon expression in Pseudomonas putida strains. We describe a molecular sensing mechanism of TodS that is activated in response to toluene. The crystal structures of the TodS Per-Arnt-Sim (PAS) 1 sensor domain (residues 43-164) and its complex with toluene (agonist) or 1,2,4-trimethylbenzene (antagonist) show a typical β2α3β3 PAS fold structure (residues 45-149), forming a hydrophobic ligand-binding site. A signal transfer region (residues 150-163) located immediately after the canonical PAS fold may be intrinsically flexible and disordered in both apo-PAS1 and antagonist-bound forms and dramatically adapt an α-helix upon toluene binding. This structural change in the signal transfer region is proposed to result in signal transmission to activate the TodS/TodT two-component signal transduction system. Site-directed mutagenesis and β-galactosidase assays using a P. putida reporter strain system verified the essential residues involved in ligand sensing and signal transfer and suggest that the Phe(46) residue acts as a ligand-specific switch.
- From the Infection and Immunity Research Center, skohtn@kribb.re.kr.
Organizational Affiliation: