Regulation of the thermoalkaliphilic F1-ATPase from Caldalkalibacillus thermarum.
Ferguson, S.A., Cook, G.M., Montgomery, M.G., Leslie, A.G., Walker, J.E.(2016) Proc Natl Acad Sci U S A 113: 10860-10865
- PubMed: 27621435
- DOI: https://doi.org/10.1073/pnas.1612035113
- Primary Citation of Related Structures:
5HKK, 5IK2 - PubMed Abstract:
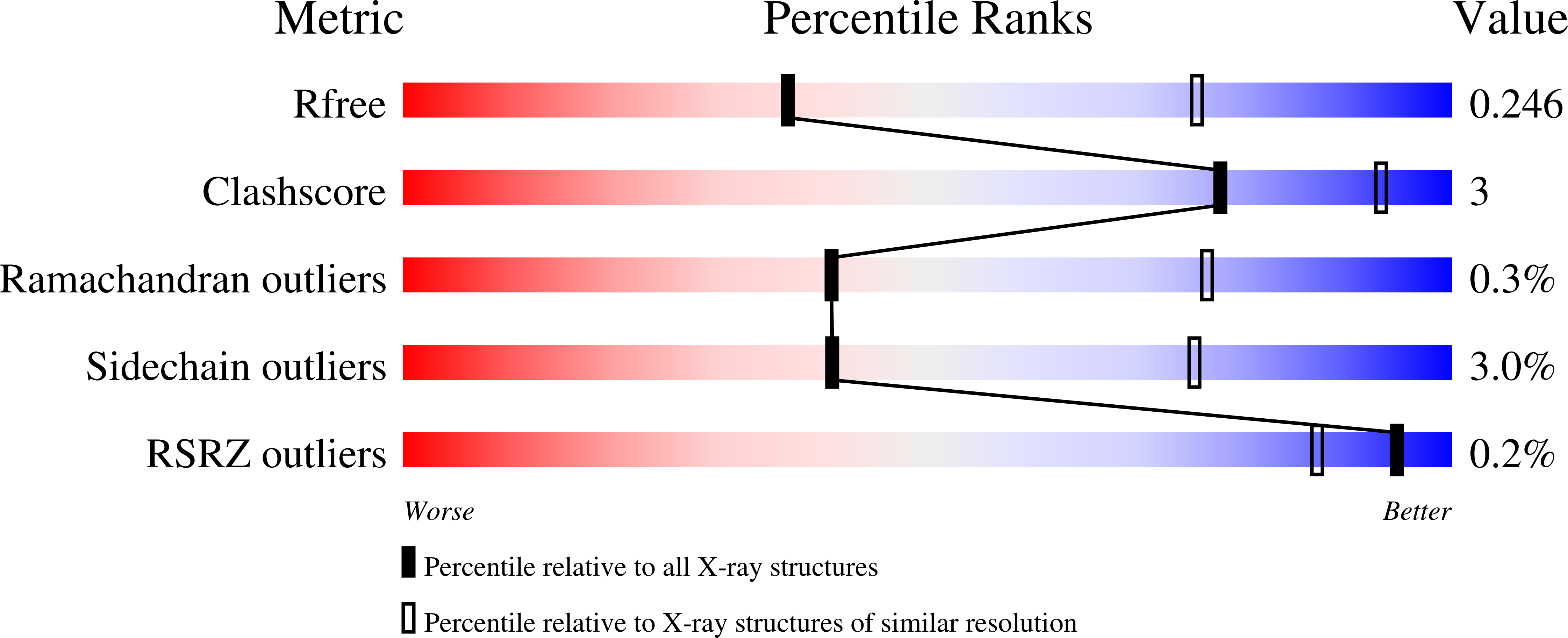
The crystal structure has been determined of the F1-catalytic domain of the F-ATPase from Caldalkalibacillus thermarum, which hydrolyzes adenosine triphosphate (ATP) poorly. It is very similar to those of active mitochondrial and bacterial F1-ATPases. In the F-ATPase from Geobacillus stearothermophilus, conformational changes in the ε-subunit are influenced by intracellular ATP concentration and membrane potential. When ATP is plentiful, the ε-subunit assumes a "down" state, with an ATP molecule bound to its two C-terminal α-helices; when ATP is scarce, the α-helices are proposed to inhibit ATP hydrolysis by assuming an "up" state, where the α-helices, devoid of ATP, enter the α3β3-catalytic region. However, in the Escherichia coli enzyme, there is no evidence that such ATP binding to the ε-subunit is mechanistically important for modulating the enzyme's hydrolytic activity. In the structure of the F1-ATPase from C. thermarum, ATP and a magnesium ion are bound to the α-helices in the down state. In a form with a mutated ε-subunit unable to bind ATP, the enzyme remains inactive and the ε-subunit is down. Therefore, neither the γ-subunit nor the regulatory ATP bound to the ε-subunit is involved in the inhibitory mechanism of this particular enzyme. The structure of the α3β3-catalytic domain is likewise closely similar to those of active F1-ATPases. However, although the βE-catalytic site is in the usual "open" conformation, it is occupied by the unique combination of an ADP molecule with no magnesium ion and a phosphate ion. These bound hydrolytic products are likely to be the basis of inhibition of ATP hydrolysis.
Organizational Affiliation:
Medical Research Council Mitochondrial Biology Unit, Cambridge Biomedical Campus, Cambridge CB2 0XY, United Kingdom;