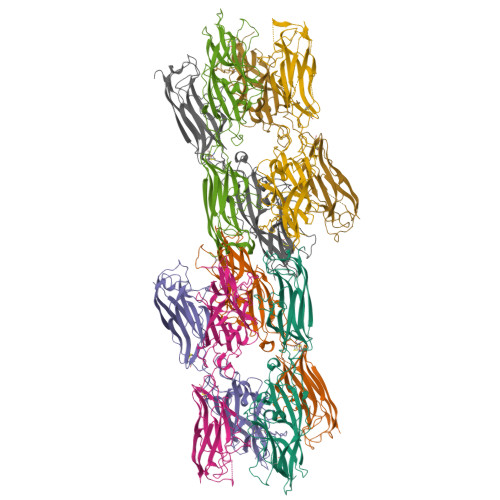
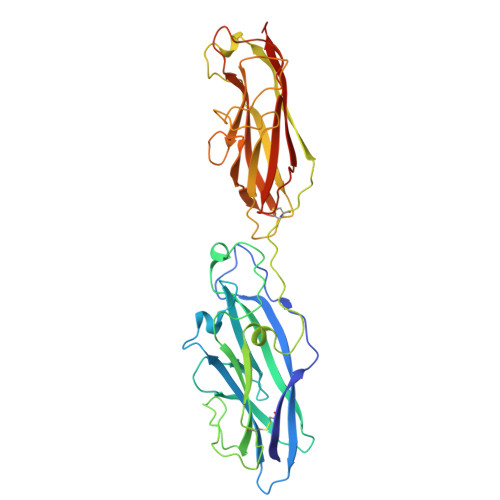
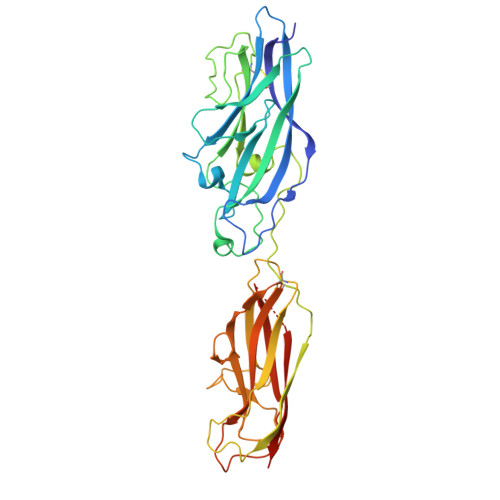
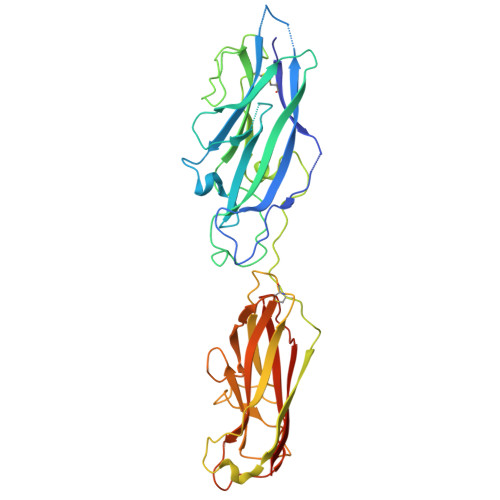
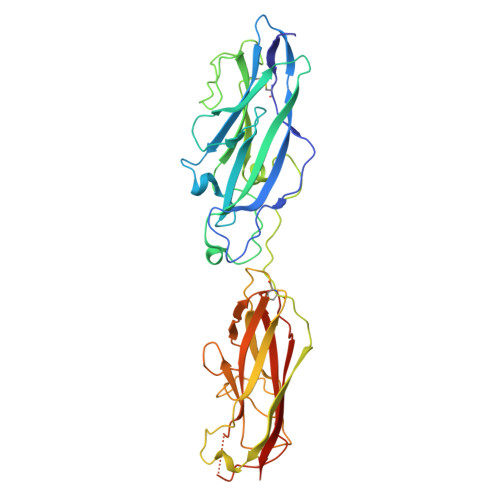
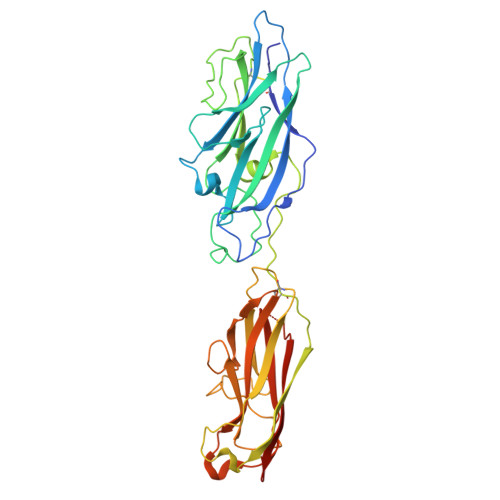
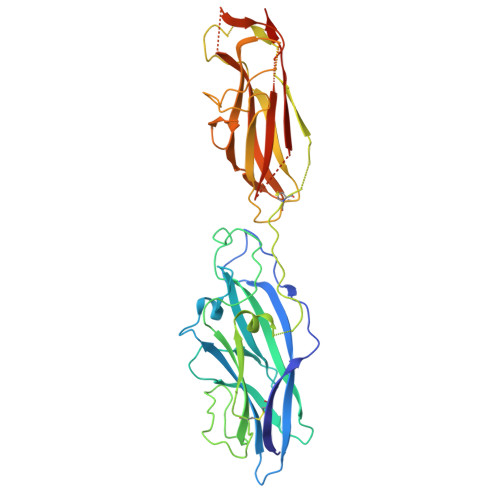
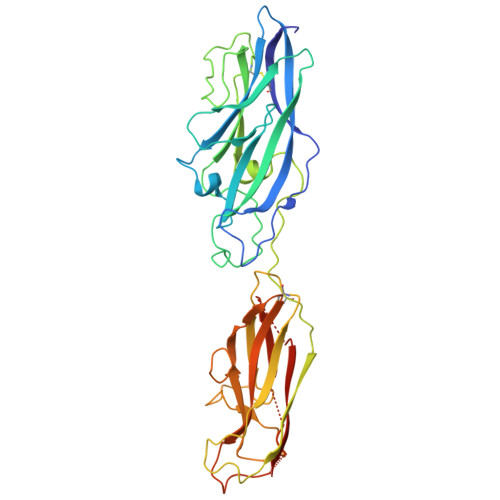
Crystal structure of the CupB6 adhesive tip from the chaperone-usher family of pili from Pseudomonas aeruginosa.
Rasheed, M., Garnett, J., Perez-Dorado, I., Muhl, D., Filloux, A., Matthews, S.(2016) Biochim Biophys Acta 1864: 1500-1505
- PubMed: 27481165
- DOI: https://doi.org/10.1016/j.bbapap.2016.07.010
- Primary Citation of Related Structures:
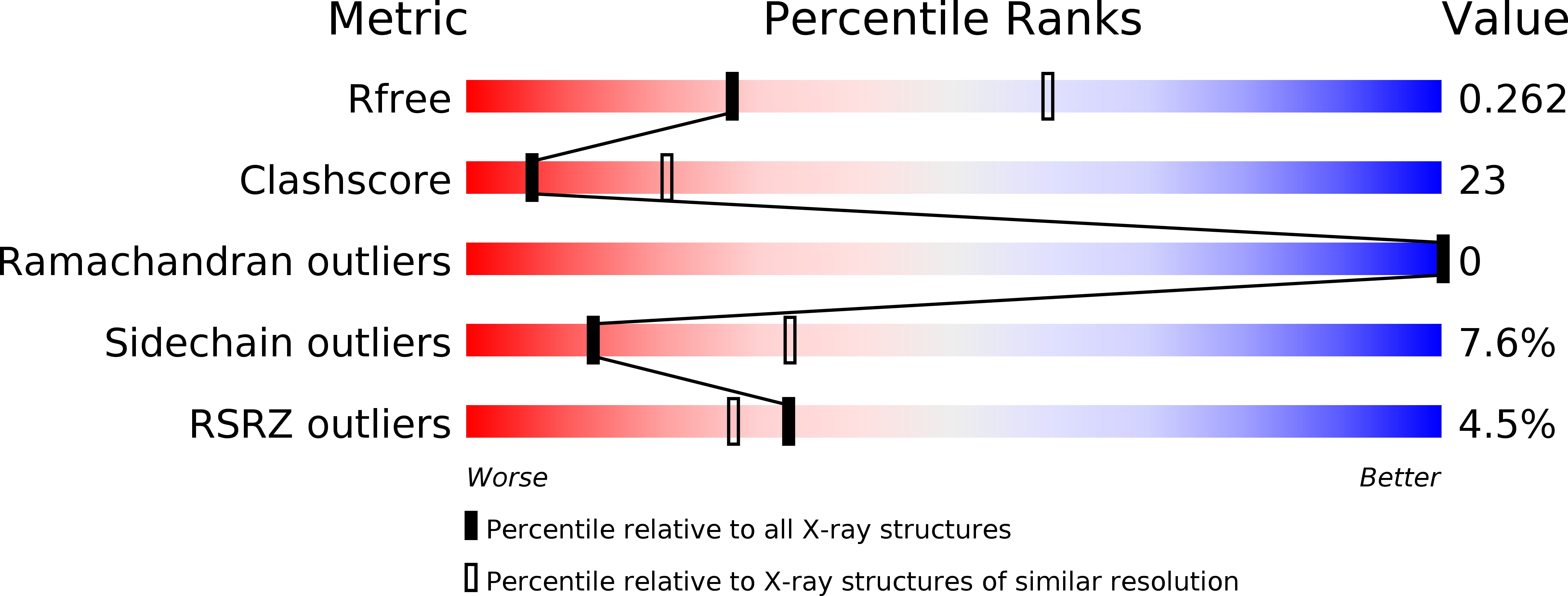
5CYL - PubMed Abstract:
Pseudomonas aeruginosa is a Gram-negative opportunistic bacterial pathogen that can cause chronic infection of the lungs of cystic fibrosis patients. Chaperone-usher systems in P. aeruginosa are known to translocate and assemble adhesive pili on the bacterial surface and contribute to biofilm formation within the host. Here, we report the crystal structure of the tip adhesion subunit CupB6 from the cupB1-6 gene cluster. The tip domain is connected to the pilus via the N-terminal donor strand from the main pilus subunit CupB1. Although the CupB6 adhesion domain bears structural features similar to other CU adhesins it displays an unusual polyproline helix adjacent to a prominent surface pocket, which are likely the site for receptor recognition.
Organizational Affiliation:
Department of Life Sciences, Imperial College London, London SW7 2AZ, United Kingdom.