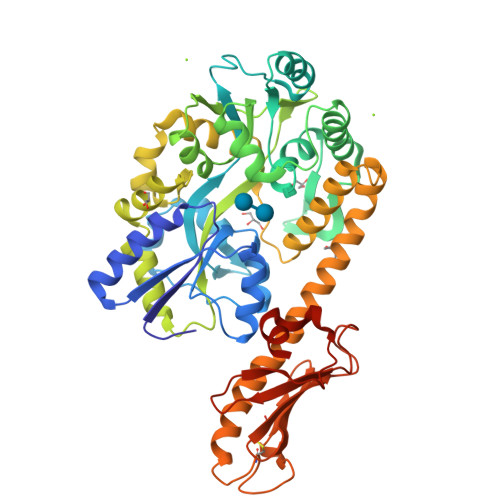
Structural Diversity in the Type IV Pili of Multidrug-resistant Acinetobacter.
Piepenbrink, K.H., Lillehoj, E., Harding, C.M., Labonte, J.W., Zuo, X., Rapp, C.A., Munson, R.S., Goldblum, S.E., Feldman, M.F., Gray, J.J., Sundberg, E.J.(2016) J Biological Chem 291: 22924-22935
- PubMed: 27634041
- DOI: https://doi.org/10.1074/jbc.M116.751099
- Primary Citation of Related Structures:
5CFV, 5IHJ - PubMed Abstract:
Acinetobacter baumannii is a Gram-negative coccobacillus found primarily in hospital settings that has recently emerged as a source of hospital-acquired infections. A. baumannii expresses a variety of virulence factors, including type IV pili, bacterial extracellular appendages often essential for attachment to host cells. Here, we report the high resolution structures of the major pilin subunit, PilA, from three Acinetobacter strains, demonstrating that A. baumannii subsets produce morphologically distinct type IV pilin glycoproteins. We examine the consequences of this heterogeneity for protein folding and assembly as well as host-cell adhesion by Acinetobacter Comparisons of genomic and structural data with pilin proteins from other species of soil gammaproteobacteria suggest that these structural differences stem from evolutionary pressure that has resulted in three distinct classes of type IVa pilins, each found in multiple species.
Organizational Affiliation:
From the Institute of Human Virology and.