Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Lee, Y., Min, C.K., Kim, T.G., Song, H.K., Lim, Y., Kim, D., Shin, K., Kang, M., Kang, J.Y., Youn, H.S., Lee, J.G., An, J.Y., Park, K.R., Lim, J.J., Kim, J.H., Kim, J.H., Park, Z.Y., Kim, Y.S., Wang, J., Kim, D.H., Eom, S.H.(2015) EMBO Rep 16: 1318-1333
- PubMed: 26341627
- DOI: https://doi.org/10.15252/embr.201540436
- Primary Citation of Related Structures:
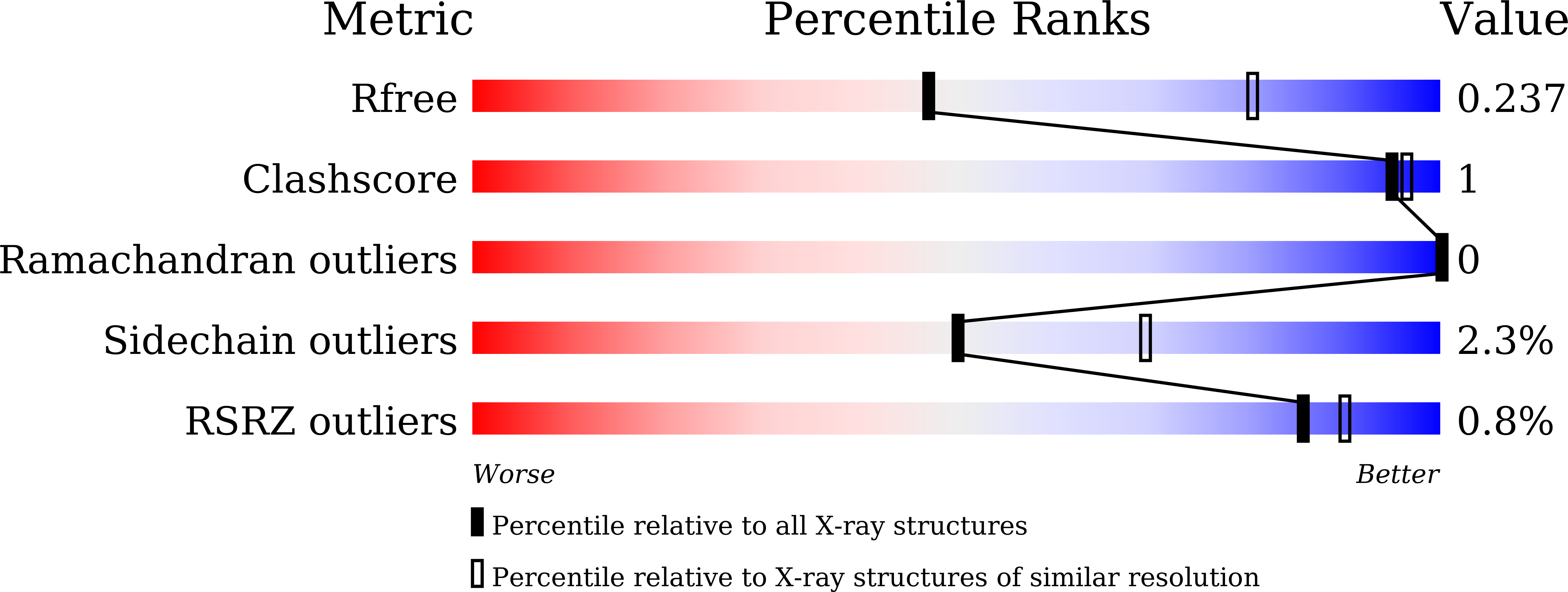
4XSJ, 4XTB, 5BZ6 - PubMed Abstract:
The mitochondrial calcium uniporter (MCU) is responsible for mitochondrial calcium uptake and homeostasis. It is also a target for the regulation of cellular anti-/pro-apoptosis and necrosis by several oncogenes and tumour suppressors. Herein, we report the crystal structure of the MCU N-terminal domain (NTD) at a resolution of 1.50 Å in a novel fold and the S92A MCU mutant at 2.75 Å resolution; the residue S92 is a predicted CaMKII phosphorylation site. The assembly of the mitochondrial calcium uniporter complex (uniplex) and the interaction with the MCU regulators such as the mitochondrial calcium uptake-1 and mitochondrial calcium uptake-2 proteins (MICU1 and MICU2) are not affected by the deletion of MCU NTD. However, the expression of the S92A mutant or a NTD deletion mutant failed to restore mitochondrial Ca(2+) uptake in a stable MCU knockdown HeLa cell line and exerted dominant-negative effects in the wild-type MCU-expressing cell line. These results suggest that the NTD of MCU is essential for the modulation of MCU function, although it does not affect the uniplex formation.
Organizational Affiliation:
School of Life Sciences, Gwangju Institute of Science and Technology (GIST), Gwangju, Korea Steitz Center for Structural Biology, Gwangju Institute of Science and Technology (GIST), Gwangju, Korea.