Assembly of Methylated Kdm1A and Chd1 Drives Androgen Receptor-Dependent Transcription and Translocation.
Metzger, E., Willmann, D., Mcmillan, J., Forne, I., Metzger, P., Gerhardt, S., Petroll, K., Von Maessenhausen, A., Urban, S., Schott, A., Espejo, A., Eberlin, A., Wohlwend, D., Schule, K.M., Schleicher, M., Perner, S., Bedford, M.T., Jung, M., Dengjel, J., Flaig, R., Imhof, A., Einsle, O., Schule, R.(2016) Nat Struct Mol Biol 23: 132
- PubMed: 26751641
- DOI: https://doi.org/10.1038/nsmb.3153
- Primary Citation of Related Structures:
5AFW - PubMed Abstract:
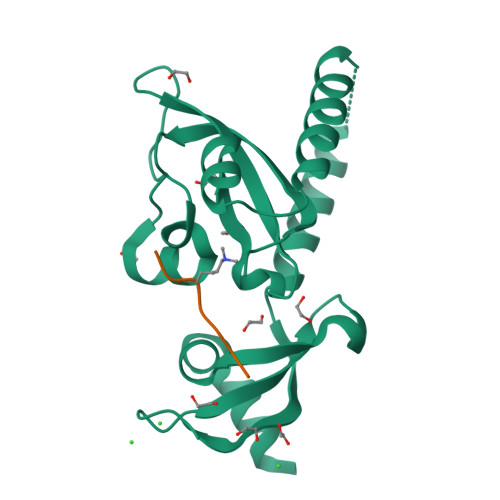
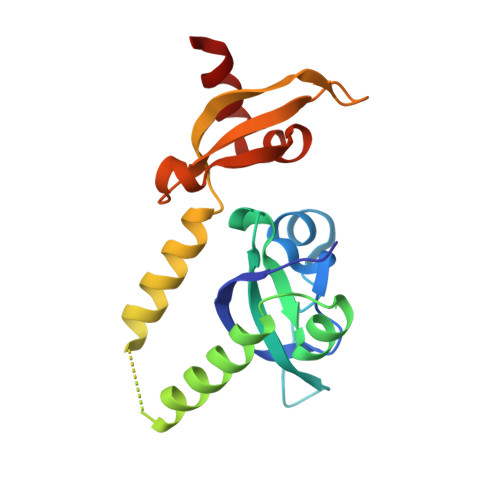
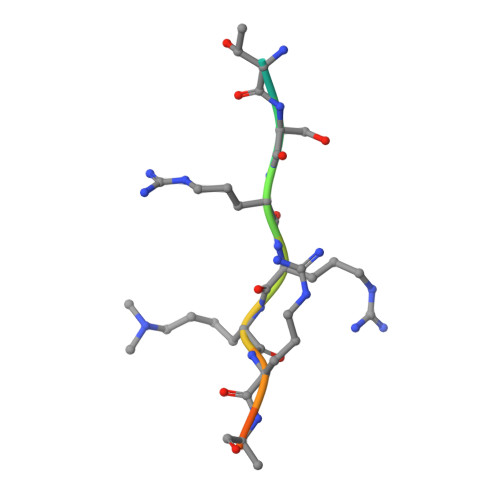
Prostate cancer evolution is driven by a combination of epigenetic and genetic alterations such as coordinated chromosomal rearrangements, termed chromoplexy. TMPRSS2-ERG gene fusions found in human prostate tumors are a hallmark of chromoplexy. TMPRSS2-ERG fusions have been linked to androgen signaling and depend on androgen receptor (AR)-coupled gene transcription. Here, we show that dimethylation of KDM1A at K114 (to form K114me2) by the histone methyltransferase EHMT2 is a key event controlling androgen-dependent gene transcription and TMPRSS2-ERG fusion. We identified CHD1 as a KDM1A K114me2 reader and characterized the KDM1A K114me2-CHD1 recognition mode by solving the cocrystal structure. Genome-wide analyses revealed chromatin colocalization of KDM1A K114me2, CHD1 and AR in prostate tumor cells. Together, our data link the assembly of methylated KDM1A and CHD1 with AR-dependent transcription and genomic translocations, thereby providing mechanistic insight into the formation of TMPRSS2-ERG gene fusions during prostate-tumor evolution.
Organizational Affiliation:
Klinik für Urologie und Zentrale Klinische Forschung, Klinikum der Universität Freiburg, Freiburg, Germany.